Generation of Human Ventral Midbrain Organoids Derived from Pluripotent Stem Cells
Edoardo Sozzi, Edoardo Sozzi, Fredrik Nilsson, Fredrik Nilsson, Janko Kajtez, Janko Kajtez, Malin Parmar, Malin Parmar, Alessandro Fiorenzano, Alessandro Fiorenzano
Abstract
Parkinson's disease (PD) is the second most common neurodegenerative disorder worldwide and is caused by the degeneration and loss of dopamine (DA) neurons in the ventral midbrain (VM). The focal and progressive degeneration of DA neurons in the VM makes PD a particularly attractive target for cell-based therapies. Human pluripotent stem cells (hPSCs) offer unprecedented opportunities to model the development and functional properties of human DA neurons in a dish. The use of human in vitro models based on hPSCs has empowered studies of VM development and provided access to neurons expressing a particular disease-specific phenotype. Currently, hPSC differentiation is most routinely carried out in monolayer cultures, which do not properly recapitulate cell-cell interactions and the structural complexity of the brain. Moreover, 2D cultures are challenging to maintain long term, as the cells tend to detach from the plate and lose their functional characteristics. This precludes the possibility of mimicking later phases of DA neurogenesis and recreating the complexity of functional neural circuitries. Here, we describe protocols showing how to maintain hPSCs in an undifferentiated state and how to then drive these hPSCs into 3D regionalized VM organoids. After long-term culture, these VM organoids exhibit mature and post-mitotic molecular features, including neuromelanin pigments similar to those released in primate VMs. We also report a protocol describing how to efficiently perform immunohistochemistry and how to detect neuromelanin-containing DA neurons in VM organoids. Together, these protocols provide a 3D in vitro platform that can be used to better understand the molecular mechanisms underlying DA neuron function and disease and may serve as a powerful tool for designing more targeted disease-modifying therapies. © 2022 The Authors. Current Protocols published by Wiley Periodicals LLC.
This article was corrected on 19 October 2024. See the end of the full text for details.
Basic Protocol 1 : Human pluripotent stem cell culture
Basic Protocol 2 : hPS cell differentiation for the generation of human ventral midbrain organoids
Basic Protocol 3 : Characterization of ventral midbrain organoids
INTRODUCTION
The brain is a complex organ composed of an enormous variety of architecturally organized cell types that give rise to intricate molecular, cellular, and biophysical processes that regulate its function and physiology (Arlotta & Pasca, 2019; Siddiqi, Kording, Parvizi, & Fox, 2022; Kelava & Lancaster, 2016). Although significant advances have been made to understand human brain development, the inaccessibility of pre- and postnatal human brain tissue severely restricts the scope of our knowledge, which, to date, has been derived mostly from studies using postmortem pathological specimens. While such studies yield data on the end-stage pathology of neurological diseases, their causes and consequences remain difficult to parse using such an approach (Krishnaswami et al., 2016; Agarwal et al., 2020). There is, therefore, a pressing need to develop novel experimental systems that can recapitulate key features of the developing and adult brain (Kim, Koo, & Knoblich, 2020; Pasca, 2018; Fiorenzano, Sozzi, Parmar, & Storm, 2021).
The ability to recreate functionally mature neurons from human pluripotent stem cells (hPSCs) holds the promise of mimicking key architectural and molecular features of brain tissue in a dish (Rifes et al., 2020; Demers et al., 2016). Currently, however, hPSC differentiation is routinely carried out in monolayer cultures, which fail to recapitulate the structural and molecular complexity of the brain. Moreover, 2D cultures are challenging to maintain long term, as the cells eventually detach from the plate and lose their functional properties. This precludes the possibility of mimicking later phases of DA neurogenesis and recreating the intricacy of neural circuitries. Recently, human brain organoids differentiated from hPSCs have emerged as an advanced human stem cell-based model capable of recapitulating the development and maturation of brain tissue in a 3D organ-like configuration in relevant physiological conditions (Lancaster et al., 2013; Quadrato et al., 2017; Kanton et al., 2019). New protocols based on extrinsic patterning factors have been developed to guide the differentiation of hPSCs toward regionalized brain organoids exhibiting a specific cell identity (Miura et al., 2020; Cederquist et al., 2019).
Dopamine (DA) neurons arise from the ventral midbrain (VM) following regionalization of the neural tube, and constitute the major source of dopamine in the mammalian central nervous system. DA neurons play a key role in the control of voluntary motor movement as well as in emotion-based behavior. The function (and dysfunction) of DA neurons has attracted great interest among the scientific community, as the degeneration of this cell population leads to motor deficits in Parkinson's disease (PD). Detailed protocols to generate human VM organoids, however, are still lacking. Moreover, modeling human VM in vitro requires the precise balance of patterning factors that lead to the generation of DA neurons exhibiting both mature molecular features and electrophysiological properties, which is not trivial (Nolbrant, Heuer, Parmar, & Kirkeby, 2017; Tiklova et al., 2020; Fiorenzano, Birtele, Wahlestedt, & Parmar, 2021; Fiorenzano et al., 2018).
Here, we report three protocols to generate and characterize human VM organoids (Fig. 1A and 1B). We first describe how to maintain hPSCs under undifferentiated conditions (Basic Protocol 1) and how to then efficiently drive these hPSCs into VM-patterned human brain organoids (Basic Protocol 2), which can give rise to mature and functional DA neurons (Fig. 1B). We then describe how to molecularly characterize these VM organoids, including the detection of DA neurons containing neuromelanin granules (Basic Protocol 3) (Fig. 1B) (Fiorenzano et al., 2021).
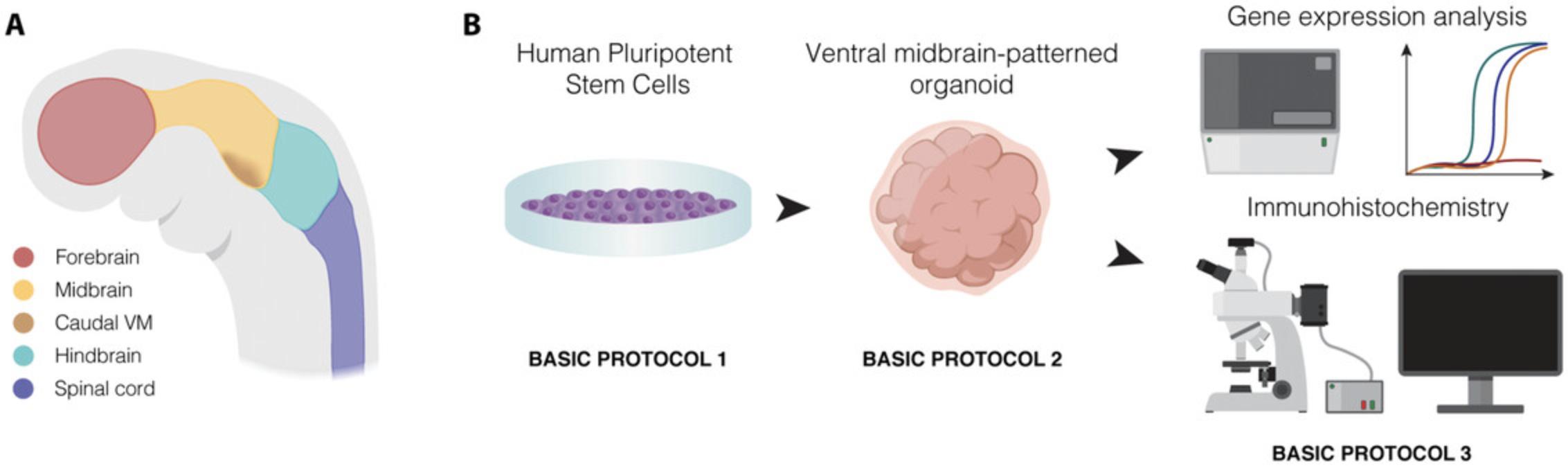
VM-patterned organoid differentiation exploits the intrinsic self-organization property of hPSCs via the addition of extrinsic patterning factors. Efficiently driving neural induction by exposing the cells to dual-SMAD inhibition factor, the neural tube ventralizing secreted factor sonic hedgehog (SHH), and glycogen synthase kinase 3 inhibitor (GSK3i) leads to finely tuned caudalized floor plate formation (Fig. 1A). This results in the efficient generation of DA progenitors within VM organoids, and their subsequent differentiation into mature DA neurons displaying electrophysiological properties and the ability to release dopamine (Fiorenzano et al., 2021). Importantly, we have previously shown that organoids patterned into a VM fate generated following this protocol give rise to mature DA neurons that are molecularly very similar to human DA neurons derived from human fetal VM (Fiorenzano et al., 2021). The recapitulation of VM patterning and the generation of mature and functional DA neurons within VM organoids offers a novel tool for molecular and functional studies of DA neuron maturation and diversity at a much greater level of detail.
Together, these protocols allow for the establishment of a valuable platform to study early phases of DA neurogenesis, but that can be maintained for a sufficiently long term to also recapitulate late stages of DA neuron differentiation, with potential implications for biomedical applications, including PD modeling and drug discovery.
Basic Protocol 1: HUMAN PLURIPOTENT STEM CELL CULTURE
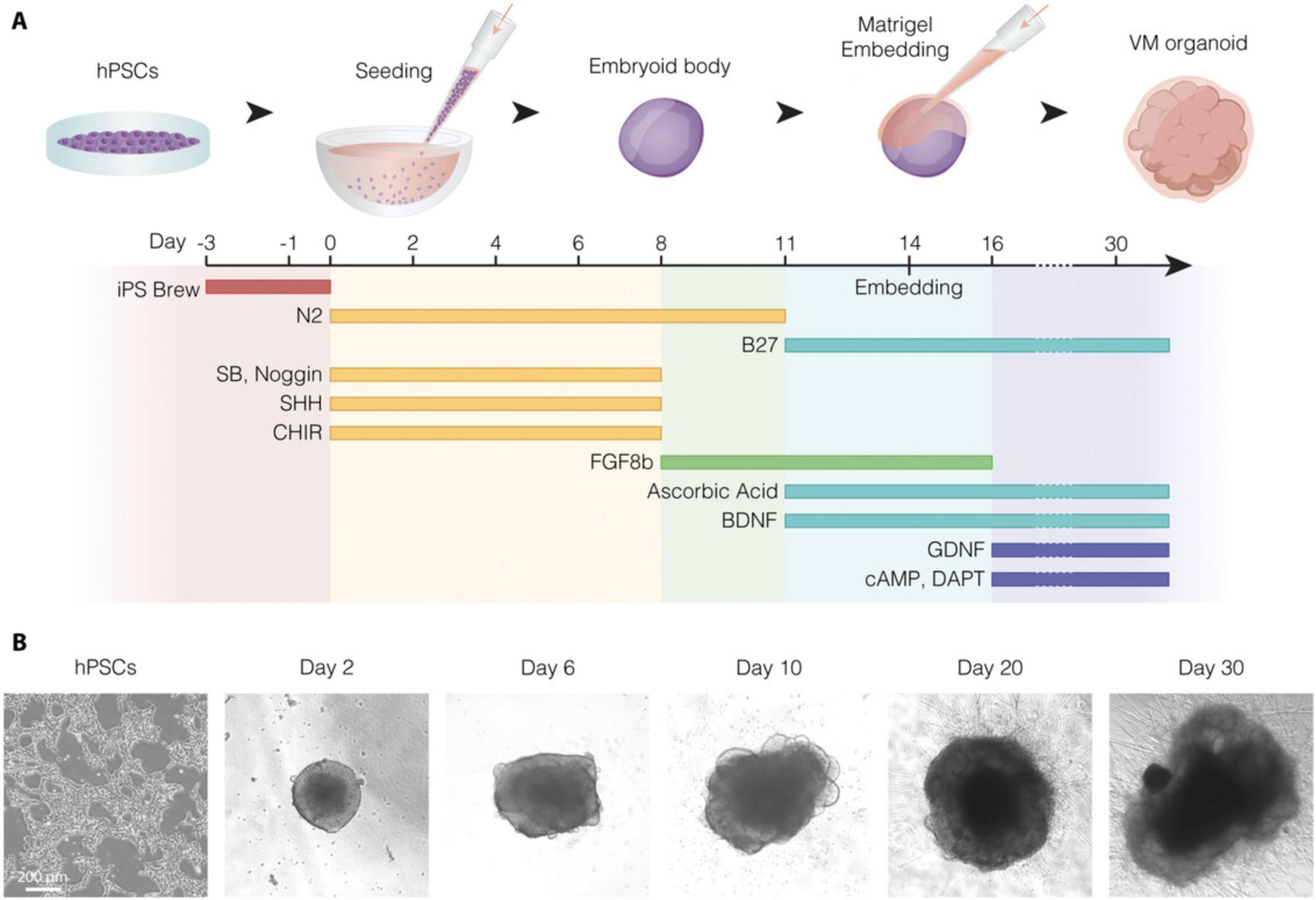
The following protocol describes how to thaw, passage, and maintain hPSCs before inducing organoid differentiation. This step is necessary to ensure that the starting population is healthy, stable, and homogeneous, without spontaneously differentiated areas. hPSC cultures should also be routinely tested for mycoplasma contamination. We recommend performing one or two passages before organoid differentiation, which can take approximately 6 to 10 days depending on the chosen stem cell line and seeding density. The protocol presented here is based on culturing hPSCs on human recombinant Lam-521-coated plates and iPS-Brew medium, which results in a homogeneous cell culture while preserving pluripotency and self-renewal properties (Fig. 2A and 2B). The protocol could also be adapted to use Matrigel coating but, in that case, we recommend monitoring the quality of hPSC cultures to be alert to the presence of areas of spontaneous differentiation, which should be manually removed.
NOTE : Users should obtain all the necessary permissions and approvals for work with hPSC lines, and follow all institutional and national guidelines.
Materials
-
hPSC line of choice. For this study, the following human embryonic stem cell lines were used: RC17 (Roslin Cells, cat. no. hPSCreg RCe021-A), H9 (WiCell, cat. no. hPSCreg WAe009A), and HS983a, HS999, and HS1001 (all from Karolinska Institute, Stockholm, Sweden).
-
Laminin-521 (Lam-521; 100 μg/ml; Biolamina, cat. no. LN-521)
-
D-PBS +Ca2+/+Mg2+ (CTS, Thermo Fisher Scientific, cat. no. A1285801)
-
DMEM/F-12 (Thermo Fisher Scientific, cat. no. 31330038)
-
Knockout serum replacement (KOSR; Thermo Fisher Scientific, cat. no. 10828010)
-
iPS-Brew XF, Stem cell culture medium (StemMACS, Miltenyi, cat. no. 130-104-368)
-
Y-27632 dihydrochloride (Rho-associated kinase [ROCK] inhibitor; StemMACS, Miltenyi, cat. no. 130-106-538)
-
EDTA (0.5 M, pH 8.0; UltraPure, Thermo Fisher Scientific, cat. no. 15575020)
-
D-PBS −Ca2+/−Mg2+ (CTS, Thermo Fisher Scientific, cat. no. A1285601)
-
Trypan blue stain 0.4% (Invitrogen, cat. no. T10282)
-
Cell culture plastic 6-well plates (Sarstedt, cat. no. 83.3920)
-
Biological safety cabinet, class II (Thermo Scientific Holten LaminAir or similar)
-
Cell culture incubator set to 37°C and 5% CO2 (Thermo Fisher Scientific, model no. 3541 DH or similar)
-
Cell culture centrifuge (for 1.5-, 15-, and 50-ml tubes; Beckman Coulter, model no. Allegra 21 or similar)
-
Countess automated cell counter (Thermo Fisher Scientific, cat. no. AMQAX1000 or AMQAX2000) or Bürker chamber (VWR, cat. no. 631-0921 or similar)
-
Pipette controller (for pipetting volumes of 1-25 ml; Corning Stripettor Ultra Pipet Controller, model no. 4099 or similar)
-
Sterile pipette tips (for pipetting volumes of 0.5-1000 µl; TipOne Pipette Tips: 1250 µl XL graduated tip, cat. no. S1112-1830; 200 µl beveled tip, cat. no. S1111-1716-C; 10 µl graduated tip, cat. no. S1111-3210 or similar)
-
Pipettes (for pipetting volumes of 0.5-1000 µl; Thermo Fisher Scientific, Finnpipette F2 variable-volume single-channel pipettes or similar)
-
Serological pipettes (sterile, non-pyrogenic; Sarstedt, 5 ml, cat. no. 86.1253.001; 10 ml, cat. no. 86.1254.001; 25 ml, cat. no. 86.1685.001)
-
Tubes (Sarstedt; 15 ml, cat. no. 62.554.502; 50 ml, cat. no. 62.547.254 or similar)
-
Freezers operating at −20°C and −80°C (Thermo Fisher Scientific, ES Series Combination Lab Refrigerator/Freezer; Thermo Fisher Scientific TSE Series −86°C, model no. 936; Panasonic Cryogenic ULT Freezer, model no. MDF-C2156VAN-PE or similar)
-
Refrigerator operating at 4°C
-
Phase-contrast inverted microscope (Olympus, model no. CKX53 or similar)
-
Automated thawing system (optional; Biocision ThawSTAR CFT2, cat. no. BCS-601)
Thawing and maintaining an hPSC line
1.Prepare a solution of Lam-521 (0.5 µg/cm2) in PBS (+Ca2+/+Mg2+) (coating solution).
2.Coat one well of a 6-well plate with 2 ml/well of the coating solution. Then, gently shake the plate to ensure a homogeneous distribution of the solution and incubate for at least 2 hr (overnight preferred) at 37°C.
3.Before starting the procedure, prepare 5 ml of wash medium (5% KOSR [v/v] in DMEM/F-12) in a 15-ml tube.
4.Thaw the cells in the automated thawing system or in a water bath at 37°C until approximately half of the medium has thawed.
5.Add 1 ml wash medium to the cryovial dropwise, making sure to collect all the cells from the walls of the vial.
6.Transfer the cells to the remaining 4 ml of wash medium (step 3) and centrifuge the tube at 400 × g for 5 min at room temperature (RT).
7.Remove the supernatant and gently knock the tube to detach the pellet from the bottom.
8.Resuspend the cells in 1 ml iPS-Brew medium supplemented with Y-27632 (ROCK inhibitor, 10 µM final concentration).
9.Count the cells with an automated counter (e.g., Countess II) or a Bürker chamber.
10.Plate cells homogeneously onto Lam-521-coated plates at a density of 15,000 cells/cm2 (approx. 144,000 cells/well for a 6-well plate). Add iPS-Brew medium supplemented with Y-27632 (ROCK inhibitor, 10 µM final concentration), to reach a final volume of 2 ml/well.
11.After 24 hr, replace the stem cell medium with 2 ml/well fresh iPS-Brew medium without the Y-27632 supplement.
12.For culture maintenance, replace the stem cell medium every day with fresh iPS-Brew medium.
Passaging of hPSC line
13.Prepare 5 ml of wash medium (5% KOSR [v/v] in DMEM/F-12) in a 15-ml tube and Lam-521-coated plates before starting, as described in steps 1-3.
14.Aspirate hPSC medium and wash the cells from step 11 once in 2 ml/well D-PBS −Ca2+/−Mg2+, to remove dead cells and debris.
15.Add 0.5 mM EDTA (50 µl/cm2, e.g., 500 µl/well for a 6-well plate) directly to the cells and incubate at 37°C for 7 min.
16.Remove the EDTA, then detach the cells by adding 1.5 ml/well wash medium. Using a 1000-µl pipette, quickly but gently triturate the colony into small clumps, without introducing bubbles.
17.Transfer all cells to the tube containing the remaining wash medium and centrifuge at 400 × g for 5 min at RT.
18.Aspirate the supernatant and detach the pellet from the bottom by gently knocking the tube.
19.Resuspend the cells in 1 ml of iPS-Brew medium supplemented with 10 µM Y-27632.
20.Count the cells with an automated counter (e.g., Countess II) or a Bürker chamber.
21.Plate cells homogeneously onto Lam-521-coated wells on iPS-Brew supplemented with 10 µM Y-27632 with the aim of having 70% confluency on the day of hPSC differentiation.
22.After 24 hr, replace the stem cell medium with fresh iPS-Brew without the Y-27632 supplement.
23.For culture maintenance, replace the stem cell medium every 1-2 days with iPS-Brew, as described above.
24.Proceed to Basic Protocol 2 when cells have reached 70% to 75% confluency.
Basic Protocol 2: hPS CELL DIFFERENTIATION FOR THE GENERATION OF HUMAN VENTRAL MIDBRAIN ORGANOIDS
This protocol describes how to guide the self-organization of the hPSCs in Basic Protocol 1 into VM-patterned organoids. The cells are first plated in conical wells to favor cell aggregation leading to the formation of embryonic bodies. Cells are then cultured under undifferentiated conditions for 3 days (day -3 to day 0). At day 0, once the cells have aggregated, the culture medium is switched to differentiation medium, which includes the addition of the following extrinsic patterning factors to efficiently drive hPSCs into VM organoids: dual-SMAD inhibitors, ventralizing agent SHH-C24II, GSK3 inhibitor (CHIR99021), and fibroblast growth factor 8b (FGF8b) (Fig. 2A and 2B and Fig. 3A-E). The proper balancing and timing of the different patterning factors used during differentiation is crucial.
This protocol has been tested on multiple hPSC and induced PSC (iPSC) lines with similar results. However, the concentration of CHIR99021 may need to be optimized, as different cell lines exhibit slightly different sensitivities to this patterning factor. CHIR99021 titration is required to identify the optimal condition for efficient VM patterning (Fig. 3F). During differentiation, when the VM organoids have grown in size, they require scaffolding to support the 3D structure. Matrigel, a natural matrix material resembling the extracellular matrix, strengthens the organization of the organoids in a 3D culture space. The organoids are embedded in Matrigel droplets at day 14 of VM differentiation to enhance organoid architecture and obtain high-order brain functions. Basic Protocol 2 can be followed to generate VM organoids containing functionally mature and pigmented DA neurons (Fig. 4), which can then be used for biomedical applications as well as for PD disease modeling.
Materials
-
Homogeneous, compact, and flat culture of hPSCs at ∼70% confluency (from Basic Protocol 1)
-
Y-27632 dihydrochloride (Rho-associated kinase [ROCK] inhibitor; StemMACS, Miltenyi, cat. no. 130-106-538)
-
D-PBS −Ca2+/−Mg2+ (CTS, Thermo Fisher Scientific, cat. no. A1285601)
-
iPS-Brew XF (StemMACS, Miltenyi, cat. no. 130-104-368)
-
DMEM/F-12 (Thermo Fisher Scientific, cat. no. 31330038)
-
Knockout serum replacement (KOSR; Thermo Fisher Scientific, cat. no. 10828010)
-
N2 base medium (see recipe)
-
B27 base medium (see recipe)
-
Accutase cell dissociation reagent (StemPro, Thermo Fisher Scientific, cat. no. A1110501)
-
SB431542 (TGF-β pathway inhibitor; StemMACS, Miltenyi, cat. no. 130-106-543)
-
Noggin (BMP inhibitor, recombinant human; Miltenyi, cat. no. 130-103-456)
-
SHH-C24II (sonic hedgehog, recombinant human; Miltenyi, cat. no. 130-095-727)
-
CHIR99021 (GSK3 inhibitor; StemMACS, Miltenyi cat. no. 130-106-539)
-
BDNF (brain-derived neurotrophic factor, recombinant human; Miltenyi, cat. no. 130-096-286)
-
GDNF (glia-derived neurotrophic factor, recombinant human; R&D Systems, cat. no. 212-GD-010)
-
FGF8b (fibroblast growth factor 8b, recombinant human; Miltenyi, cat. no. 130-095-740)
-
cAMP (dibutyryl-cyclic AMP; Sigma-Aldrich, cat. no. D0627)
-
DAPT (Tocris Bioscience, cat. no. 2634)
-
l-ascorbic acid (vitamin C; Sigma-Aldrich, cat. no. A4403)
-
Matrigel matrix basement membrane (Corning, cat. no. 354234)
-
Trypan blue stain 0.4% (if an automated cell counter is used; Invitrogen, cat. no. T10282)
-
Biological safety cabinet, class II (Thermo Scientific Holten LaminAir or similar)
-
Cell culture centrifuge (for 1.5-, 15-, and 50-ml tubes; Beckman Coulter, model no. Allegra 21 or similar)
-
Cell culture incubator set to 37°C and 5% CO2 (Thermo Fisher Scientific, model no. 3541 DH or similar)
-
Countess automated cell counter (Thermo Fisher Scientific, cat. no. AMQAX1000 or AMQAX2000) or Bürker chamber (VWR, cat. no. 631-0921 or similar)
-
Pipette controller (for pipetting volumes of 1-25 ml; Corning Stripettor Ultra Pipet Controller, model no. 4099 or similar)
-
Sterile pipette tips (for pipetting volumes of 0.5-1000 µl; TipOne Pipette Tips: 1250 µl XL graduated tip, cat. no. S1112-1830; 200 µl beveled tip, cat. no. S1111-1716-C; 10 µl graduated tip, cat. no. S1111-3210 or similar)
-
Pipettes (for pipetting volumes of 0.5-1000 µl; Thermo Fisher Scientific, Finnpipette F2 variable-volume single-channel pipettes or similar)
-
Electronic Multi-Dispenser Pipette (Multipette® E3×, Eppendorf)
-
Serological pipettes (sterile, non-pyrogenic; Sarstedt, 5 ml, cat. no. 86.1253.001; 10 ml, cat. no. 86.1254.001; 25 ml, cat. no. 86.1685.001)
-
Tubes (Sarstedt; 15 ml, cat. no. 62.554.502; 50 ml, cat. no. 62.547.254 or similar)
-
Freezers operating at −20°C and −80°C (Thermo Fisher Scientific, ES Series Combination Lab Refrigerator/Freezer; Thermo Fisher Scientific TSE Series −86°C, model no. 936; Panasonic Cryogenic ULT Freezer, model no. MDF-C2156VAN-PE or similar)
-
Refrigerator operating at 4°C
-
Phase-contrast inverted microscope (Olympus, model no. CKX53 or similar)
-
Ultra-low attachment 96-well plate (round bottom; Corning Costar, cat. no. 7007)
-
Ultra-low attachment 6-well plate (Corning Costar, cat. no. 3471)
-
Parafilm (Sigma-Aldrich, cat. no. P7793 or similar)
Cell seeding (day –3)
1.Before differentiation, make sure that the hPSC culture from Basic Protocol 1 looks homogeneous and flat, without areas undergoing spontaneous differentiation.
2.Prepare 5 ml wash medium (5% KOSR [v/v] in DMEM/F-12) in a 15-ml tube.
3.Aspirate hPSC medium and wash the cells once in 2 ml/well of D-PBS −Ca2+/−Mg2+.
4.Incubate the cell culture with Accutase solution (50 µl/cm2, e.g., 500 µl/well for a 6-well plate) at 37°C for 5 min.
5.Without removing the Accutase solution, gently add 1.5 ml wash medium to each well to detach the cells, using a 1000 µl pipette to mechanically obtain small cell clumps and single cells.
6.Transfer cell suspensions to the tube from step 2 and spin down at 400 × g for 5 min at RT.
7.Aspirate the supernatant and resuspend the pellet in 1 ml iPS-Brew supplemented with Y-27632 (ROCK inhibitor, 10 µM).
8.Count the cells with an automated counter (e.g., Countess II) or a Bürker chamber.
9.Dilute the cell suspension to a concentration of 160,000 cells/ml in the same medium.
10.Plate 8000 cells/well (e.g., 50 µl/well) in all wells of one ultra-low attachment 96-well plate, mixing the suspension from time to time to ensure an equal distribution of cells in all wells.
11.Gently add 200 µl/well of iPS-Brew supplemented with 10 µM Y-27632 (total volume 250 µl/well) and leave the plate undisturbed in the incubator.
12.After 48 hr (day –1), replace 150 µl of medium with 200 µl of fresh iPS-Brew without the Y-27632 supplement.
13.Wait 24 hr before proceeding to the next step.
Embryoid body formation and patterning (days 0 to 14)
14.Prepare N2 base medium supplemented with 10 µM SB431542, 150 ng/ml Noggin, 1.5 µM CHIR99021, and 400 ng/ml SHH-C24II.
15.Gently replace 200 µl/well medium with the N2 medium.
16.Replace 250 µl/well with N2 medium and fresh supplements (as above) on days 2, 4, and 6.
17.On day 8, replace 250 µl/well with N2 medium supplemented with fresh 100 ng/ml FGF8b.
18.On day 11, replace 250 µl/well with B27 medium supplemented with fresh 100 ng/ml FGF8b, 20 ng/ml BDNF, and 200 µM ascorbic acid.
Matrigel embedding (days 13 and 14) and terminal differentiation (day 16 onwards)
19.On day 13, before starting the embedding procedure, let the Matrigel thaw undisturbed overnight at 2°-8°C in a bucket of ice.
20.On day 14, take a support surface with holes arranged horizontally and vertically (e.g., a 96-well plate), coat the surface with a Parafilm layer, and apply finger pressure to generate dimples. Sterilize the dimples with UV/ethanol.
21.Using a cut P1000 tip, transfer the embryoid bodies from the corresponding well to individual dimples in a small drop of medium.
22.With a P200 tip, remove as much medium as possible from the surroundings of each 3D culture.
23.Evenly cover each embryoid body with a droplet of Matrigel (∼30 µl/organoid).
24.Incubate the plate at 37°C for 30 min.
25.After embedding, transfer the organoids to an ultra-low attachment 6-well plate, at approximately 10 organoids/well. To facilitate the transfer, detach the Parafilm from the support and wash out the embedded organoids with 300 µl/dimple of B27 base medium supplemented with fresh 20 ng/ml BDNF, 10 ng/ml GDNF, 200 µM ascorbic acid, 500 µM cAMP, and 1 µM DAPT. Once all organoids are transferred to the corresponding wells, add medium to reach a final volume of 4 ml/well.
26.For terminal differentiation of the culture, replace 4 ml/well of the medium 2 to 3 times a week for 2 to 4 months (depending on the experimental design) with B27 base medium supplemented with fresh 20 ng/ml BDNF, 10 ng/ml GDNF, 200 µM ascorbic acid, 500 µM cAMP, and 1 µM DAPT. Depending on the experimental design, the organoids can now be analyzed at different time points with the technique of choice. To perform IHF/IHC, proceed to Basic Protocol 3.
| Gene name | Full gene name | Primer sequence (fwd/rev, 5′-3′) |
|---|---|---|
| ACTB | Actin beta | CCTTGCACATGCCGGAGGCACAGAGCCTCGCCTT |
| GAPDH | Glyceraldehyde-3-phosphate dehydrogenase | TTGAGGTCAATGAAGGGGTCGAAGGTGAAGGTCGGAGTCA |
| NANOG | Homeobox Transcription Factor Nanog | TTGGGACTGGTGGAAGAATCGATTTGTGGGCCTGAAGAAA |
| OCT4 (POU5F1) | Octamer-Binding Protein 4 | TCTCCAGGTTGCCTCTCACTGTGGAGGAAGCTGACAACAA |
| SOX17 | SRY-Box Transcription Factor 17 | CCAGACCGCGACAGGCCAGAACAGTGAGGCACTGAGATGCCCCGAG |
| FOXA2 | Forkhead box A2 | CCGTTCTCCATCAACAACCTGGGGTAGTGCATCACCTGTT |
| CORIN | Corin, serine peptidase | CATATCTCCATCGCCTCAGTTGGGCAGGAGTCCATGACTGT |
| LMX1B | LIM homeobox transcription factor beta | CTTAACCAGCCTCAGCGACTTCAGGAGGCGAAGTAGGAAC |
| EN1 | Engrailed homeobox 1 | CGTGGCTTACTCCCCATTTATCTCGCTGTCTCTCCCTCTC |
| NURR1 (NR4A2) | Nuclear Receptor Subfamily 4 Group A Member 2 | CAGGCGTTTTCGAGGAAATGAGACGCGGAGAACTCCTAA |
| TH | Tyrosine Hydroxylase | CGGGCTTCTCGGACCAGGTGTACTCCTCGGCGGTGTACTCCACA |
| FOXG1 | Forkhead box G1 | TGGCCCATGTCGCCCTTCCTGCCGACGTGGTGCCGTTGTA |
| HOXA2 | Homeobox A2 | CGTCGCTCGCTGAGTGCCTGTGTCGAGTGTGAAAGCGTCGAGG |
| SHH | Sonic Hedgehog Signaling Molecule | CCAATTACAACCCCGACATCAGTTTCACTCCTGGCCACTG |
| MASH1 (ASCL1) | Achaete-Scute Family BHLH Transcription Factor 1 | CTAAAGATGCAGGTTGTGCGGGAGCTTCTCGACTTCACCA |
| EN2 | Engrailed Homeobox 2 | CCTCCTGCTCCTCCTTTCTTGACGCAGACGATGTATGCAC |
| OTX1 | Orthodenticle homeobox 1 | TATAAGGACCAAGCCTCATGGCTTCTCCTCTTTCATTCCTGGGC |
| TBR2 (EOMES) | Eomesodermin | GCGAGAGAACCGTGCCACAGACGCCACCTCTTCGCTCTGTTGGG |
| MAP2 | Microtubule Associated Protein 2 | CCGTGTGGACCATGGGGCTGGTCGTCGGGGTGATGCCACG |
| NEUROG2 | Neurogenin 2 | ATCCGAGCAGCACTAACACGGCACAGGCCAAAGTCACAG |
Basic Protocol 3: CHARACTERIZATION OF VENTRAL MIDBRAIN ORGANOIDS
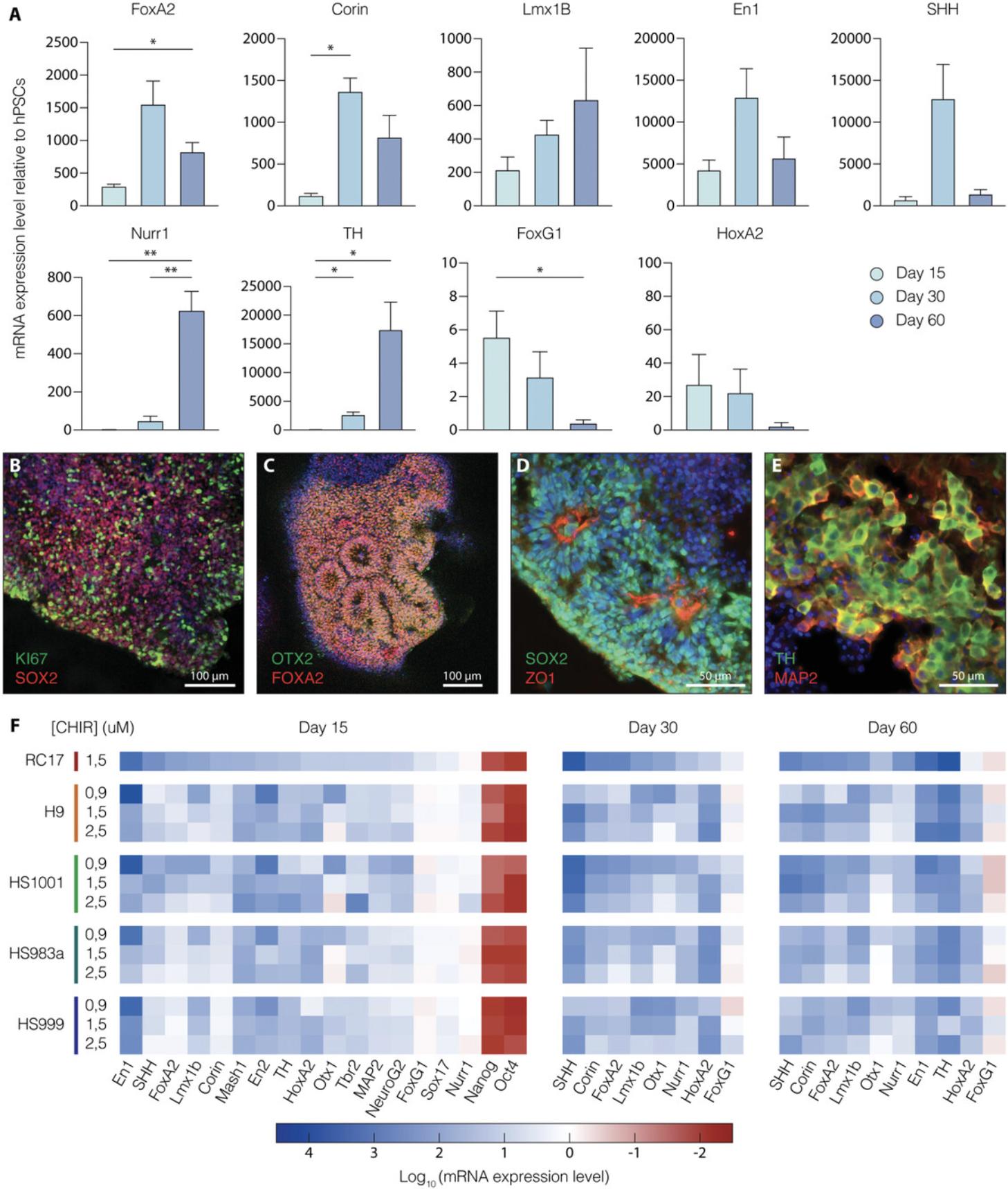
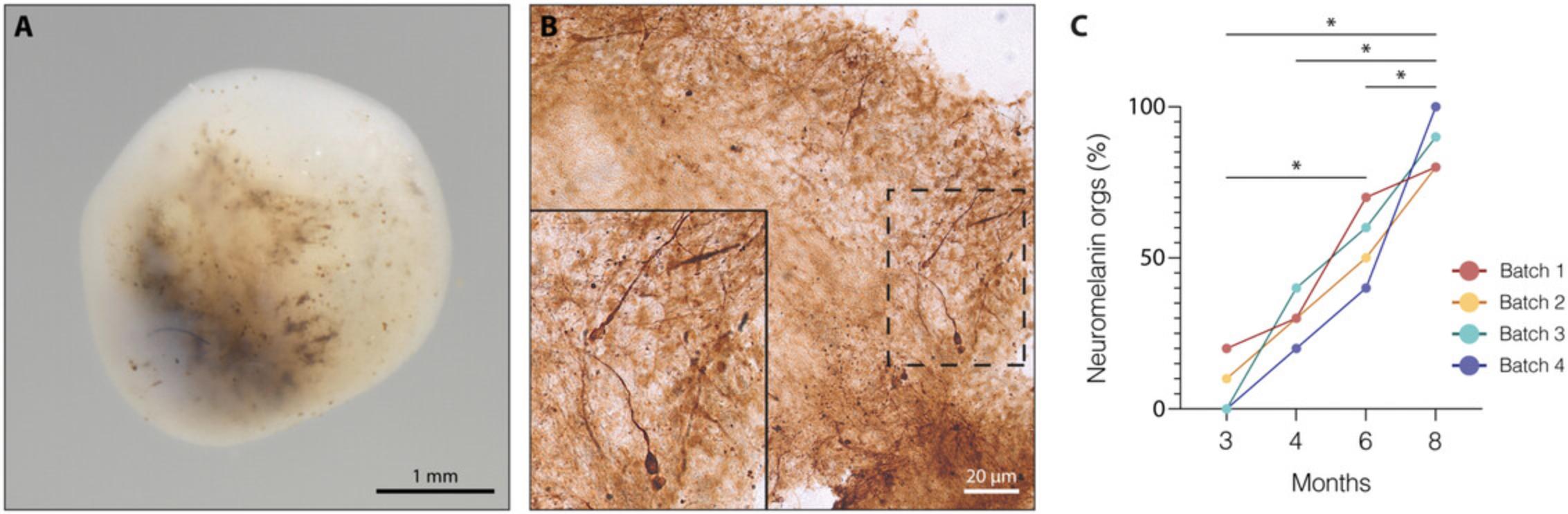
During the differentiation of human VM organoids, cells in 3D space start exhibiting distinct molecular and architectural features. The presence of characteristic cell structures with apical-basal polarity and the formation of distinct cell layers is commonly analyzed by immunofluorescence staining using specific antibodies for marker proteins. The expression of immature and post-mitotic DA neuronal markers, including FOXA2, LMX1A, OTX2, and TH, detected by immunohistochemistry, is also specifically used to analyze VM differentiation efficiency. Specifically, maturation to A9-like DA neurons, which consist of pigmented neurons located in substantia nigra pars compacta (SNpc) in primate VM (Bjorklund & Dunnett, 2007), can be detected using Fontana-Masson staining in combination with tyrosine hydroxylase (TH) immunohistochemistry, which reveals the presence of dark neuromelanin granules (Figs. 3E and 4B).
Here, we describe how to molecularly characterize VM organoids generated from Basic Protocol 2 at the protein level. We report a step-by-step protocol for embedding and sectioning VM organoids to guarantee an efficient immunohistochemical analysis, including detection of neuromelanin pigmented DA neurons after long-term culture. This protocol is based on Fiorenzano et al. (2021).
Materials
- Ventral midbrain patterned organoids generated from Basic Protocol 2, at the desired time point.
- Paraformaldehyde (PFA; Merck Millipore, cat. no. 1040051000)
- Sucrose (Fisher Chemical, cat. no. S/8600/60)
- OCT cryo embedding compound (HistoLab, cat. no 45830)
- D-PBS −Ca2+/−Mg2+ (CTS, Thermo Fisher Scientific, cat. no. A1285601)
- Donkey serum (Merck Millipore, cat. no. S30-100ML)
- Triton X-100 (Fisher Scientific, cat. no. 10254640)
- Primary antibodies (Table 2)
- DAPI (Sigma-Aldrich, cat. no. D9542)
- Secondary conjugated antibodies:
- Alexa Fluor 488 AffiniPure Donkey Anti-Rabbit IgG (H+L) (Jackson ImmunoResearch Laboratories, cat. no. 711-545-152)
- Alexa Fluor 488 AffiniPure Donkey Anti-Goat IgG (H+L) (Jackson ImmunoResearch Laboratories, cat. no. 705-545-003)
- Cy2 AffiniPure Donkey Anti-Rabbit IgG (H+L) (Jackson ImmunoResearch Laboratories, cat. no. 711-225-152)
- Cy2 AffiniPure Donkey Anti-Mouse IgG (H+L) (Jackson ImmunoResearch Laboratories, cat. no. 715-545-150)
- Cy3 AffiniPure Donkey Anti-Rabbit IgG (H+L) (Jackson ImmunoResearch Laboratories, cat. no. 711-165-152)
- Cy3 AffiniPure Donkey Anti-Chicken IgG (H+L) (Jackson ImmunoResearch Laboratories, cat. no. 703-165-155)
- Cy3 AffiniPure Donkey Anti-Mouse IgG (H+L) (Jackson ImmunoResearch Laboratories, cat. no. 715-165-150)
| Antigen | Species | Brand (cat. no) | Dilution |
|---|---|---|---|
| KI67 | Mouse | Novocastra (ACK02) | 1:500 |
| SOX2 | Rabbit | Millipore (AB5603) | 1:400 |
| FOXA2 | Mouse | Santa Cruz (sc-101060) | 1:1000 |
| OTX2 | Goat | R&D Systems (AF1979) | 1:2000 |
| TH | Rabbit | Merck Millipore (AB152) | 1:1000 |
| MAP2 | Chicken | Abcam (ab5392) | 1:2000 |
| ZO1 | Mouse | Invitro (339100) | 1:300 |
- Neuromelanin staining
- Staining jars (Epredia E94 or similar)
- Fontana-Masson kit (Atom Scientific, cat. no. RRSK 12-100)
- Cell culture plastic 24-well plates (Sarstedt, cat. no. 83.3922 or similar)
- Cryostat (Fisher Scientific CryoStar NX70, or similar)
- Fluorescence microscope (Leica, model no. DMI6000 B or similar)
- Confocal laser scanning microscope (Leica, model no. TCS SP8 or similar)
- High-resolution bright-field microscope (Olympus, model no. AX70 or similar)
- Disposable base molds (Fisherbrand, Fisher Scientific, cat. no. 93019849)
- Microscope slides (Epredia SuperFrost Plus, Fisher Scientific, cat. no. J1800AMNZ or similar)
Organoid cryosectioning and immunofluorescence
1.Transfer 3 to 6 VM organoids at the desired time point (from Basic Protocol 2) to a single well of a 24-well plate, remove any residual medium, and fix them with 500 µl/well of 4% v/v paraformaldehyde for 5 hr at RT.
2.Perform three 5-min washes with 1 ml/well of 1× PBS.
3.Remove the PBS and add 2 ml of 30% w/v sucrose to each well and leave overnight at 4°C on a shaker.
4.Replace sucrose solution with 2 ml of 1:1 (by volume) OCT:30% w/v sucrose mixture for 6 hr at 4°C on an orbital shaker.
5.Use a 5-ml pipette to transfer VM organoids to a cryomold and fill with OCT.
6.Immediately transfer the cryomold to dry ice and leave it to freeze for 5 min.
7.Using a cryostat, section the organoids embedded in OCT at 20 μm and then transfer the sections onto glass slides with the help of a brush.
8.Wash the slides in 1× PBS three times for 5 min each.
9.Fix the slides in 4% v/v paraformaldehyde for 10 min at RT and wash again three times with 1× PBS, as in step 8.
10.Incubate each slide with 500 µl of 0.3% v/v Triton X-100 and 5% v/v donkey serum in 1× PBS for at least 1 hr at RT.
11.Prepare primary antibodies solutions by diluting them in the same solution as step 10.
12.Incubate each slide with 300 µl of primary antibody solution overnight at 4°C.
13.The next day, wash the sections in 1× PBS three times, for 5 min each.
14.After incubation with primary antibodies, incubate the sections for at least 1 hr with 300 µl/slide of the appropriate secondary antibodies (Alexa Fluor 488, 594, and 647 used at 1:400 in the same solution as in step 10) and then mount with coverslip and 500 µl PVA-DABCO containing DAPI (1:1000).
15.Analyze the immunocytochemically stained VM organoid sections using a fluorescence microscope or a confocal laser scanning microscope to estimate the expression pattern of early and late DA neuron markers. See Figure 3B-E.
Immunohistochemistry for neuromelanin detection
The following protocol is based on the Fontana-Masson method to visualize melanin in cells, as well as argentaffin cell granules and lipofuscins, which takes advantage of a silver-reducing technique. All reagents are included in the Fontana-Masson staining kit listed in Materials. This protocol is also compatible with DAB pre-stained tissue. All steps are performed in staining jars; make sure that all sections are completely covered with solution.
16.Prepare the ammoniacal silver solution by adding 33% ammonia solution drop by drop to 20 ml of 10% aqueous silver nitrate solution in a glass flask. Constantly agitate the flask to favor the dissolution of the formed precipitate. Stop adding the ammonia solution once a faint opalescence is obtained and the precipitate is dissolved.
17.After titration, add 20 ml distilled water.
18.Hydrate the sections by rinsing them in tap water.
19.Wash the sections three times for 1 min each in distilled water.
20.Transfer the slides to a clean jar filled with the ammoniacal silver solution from step 17, cover with aluminum foil, and incubate in an oven at 56°C for 30 to 40 min.
21.Gently wash the slides at least three times for 1 min each in distilled water.
22.Immerse the slides in 5% aqueous sodium thiosulphate solution for 1 min.
23.Wash well in running tap water for 3 min and rinse in distilled water.
24.Counterstain with 0.1% neutral red solution for 1 min.
25.Rinse in distilled water.
26.Rapidly dehydrate the sections by rinsing them in absolute ethanol three times, for 1 min each.
27.Clear the sections by immersion in fresh xylene three times, for 1 min each, and place under a coverslip with DPX mountant.
28.Analyze the immunohistochemically stained VM organoid section using a bright-field microscope to quantify the number of neuromelanin+ organoids (Fig. 4).
REAGENTS AND SOLUTIONS
N2 base medium
| Dilution | Final conc. | Amount | Vendor | Cat. no. | |
|
DMEM/F-12 (no glutamine) |
1:2 | 0.5× | 19.4 ml | Thermo Fisher | 21331020 |
| Neurobasal Plus | 1:2 | 0.5× | 19.4 ml | Thermo Fisher | A3582901 |
| N2 supplement | 1:100 | 1× | 400 µl | Thermo Fisher | 17502048 |
| l-Glutamine | 1:100 | 2 mM | 400 µl | Thermo Fisher | 25030081 |
| Penicillin-Streptomycin | 1:500 | 20 U/ml | 80 µl | Thermo Fisher | 15140122 |
| MEM Non-essential amino acids solution | 1:100 | 1× | 400 µl | Thermo Fisher | 11140050 |
| 2-Mercaptoethanol | 1:1000 | 50 μM | 40 µl | Thermo Fisher | 31350010 |
| Total | 40 ml |
B27 base medium
| Dilution | Final conc. | Amount | Vendor | Cat. no. | |
| Neurobasal Plus | – | 1× | 38.4 ml | Thermo Fisher | A3582901 |
| B27 supplement (without vitamin A) | 1:50 | 1× | 800 µl | Thermo Fisher | 12587010 |
| l-Glutamine | 1:100 | 2 mM | 400 µl | Thermo Fisher | 25030081 |
| Penicillin-Streptomycin | 1:500 | 20 U/ml | 80 µl | Thermo Fisher | 15140122 |
| MEM Non-essential amino acids solution | 1:100 | 1× | 400 µl | Thermo Fisher | 11140050 |
| 2-Mercaptoethanol | 1:1000 | 50 μM | 40 µl | Thermo Fisher | 31350010 |
| Total | 40 ml |
COMMENTARY
Background information
Monolayer 2D culture systems laid the foundation for understanding the early developmental stages of DA neurons, enabling the development and optimization of more efficient differentiation protocols. This knowledge has now been transferred to 3D models, which are able to mimic the architectural organization and functional properties of DA neurons in a more physiologically relevant condition. Indeed, in the last decade, human brain organoids have emerged as a useful tool for recapitulating key aspects of mature DA neuron tissue. By exploiting specific extrinsic patterning factors, hPSCs can now be differentiated, in a 3D space, toward a VM fate, resulting in regionalized human VM organoids. These VM 3D structures reproduce later stages of midbrain development in a dish and can result in the generation of functionally mature DA neurons with post-mitotic molecular features as well as electrophysiological activity. Additionally, distinct DA neuron subtypes also emerge within these VM organoids, mimicking human dopamine diversity in vitro. Neuromelanin-pigmented DA neurons, similar to A9 populations in SNpc in VM primates, can be detected in long-term 3D culture (Jo et al., 2016; Fiorenzano et al., 2021). Given their ability to generate functionally mature DA neurons releasing neuromelanin, VM organoids also represent a valid tool for PD modeling using iPSCs from PD patients. PD is considered one of the most complex neurological disorders, as it results from the intricate interaction of genetic, epigenetic, and environmental factors that make the study of this disease extremely challenging in both animal models and 2D culture.
Here, we describe a set of protocols from hPSC culture to the generation and characterization of VM organoids that can then be used to study the mechanisms underlying the development and maturation of DA neurons and to model pathological phenotypes by recapitulating the molecular hallmarks observed in PD patients.
Critical Parameters
The protocol described here can be adopted in different laboratories to differentiate VM organoids using different hPSC as well as iPSC lines. High-quality hPSC culture prior to differentiation is critical. For this, i) hPSC colonies should be maintained at around 70% to 80% confluence; ii) no areas undergoing spontaneous differentiation should be present; and iii) hPSCs should be routinely tested to ensure they are free from mycoplasma infection, using a mycoplasma PCR detection kit (Thermo Fisher Scientific, cat. no. 4460626 or similar).
Accurate titration of patterning factors during the differentiation of VM organoids is also crucial. To avoid repeated freeze-thawing that could alter the efficiency of differentiation, growth factors should be stored in working aliquots at −20°C. It is also important to bear in mind that different hPSCs may require different concentrations of patterning factors during differentiation. For example, the concentration of CHIR99021 proved to be particularly critical in our assays. For this reason, we suggest adapting our protocol to differentiate different cell lines by testing different CHIR99021 concentrations, to avoid contamination of non-VM progenitors. In that case, we suggest including two negative controls: i) differentiation without patterning factors and ii) cerebral organoid differentiation as a negative control of neuromelanin release (Table 3).
| Problem | Possible cause | Solution |
|---|---|---|
| No embryoid bodies are formed by day 2 | Number of cells too low |
Check cell counting Make sure Y-27632 (ROCK inhibitor) was added |
| Different-sized organoids at day 2 | Plating a variable of cell number in each microwell | Use automatic pipetting |
| Yellow medium at day 15 | High organoid growth at this stage | Add larger volume of medium (6 ml/well) |
| Poor expression of LMX1+/FOXA2+ as assessed by RT-qPCR | Organoids have assumed a too rostral identity expressing FOXG1 | Increase CHIR99021 concentration to 2.5 μM in order to achieve a more caudal patterning of VM organoids |
| Absence of bright buds and layer generation on days 10-20 |
Medium composition Working aliquot was subjected to multiple freeze/thaw cycles |
Thaw new reagents and prepare fresh CHIR99021 aliquots |
| TH expression is restricted to the outer region of VM organoids. | The collected sections were derived from the organoid inner core | Collect internal and external organoid sections on the same slide during cryosectioning |
| Weak immunohistochemical expression of DA neuron markers | Organoids not properly fixed in 4% paraformaldehyde (Basic Protocol 3 step 1) | Increase paraformaldehyde concentration to 6% in Basic Protocol 3 step 9 |
| Absence of neuromelanin release | Medium composition | Organoids may show either a too rostral (forebrain) or a too caudal (hindbrain) patterning |
During VM organoid generation, we recommend using the same batch of Matrigel during embedding of the organoids to avoid variability in differentiation efficiency and in cellular composition. An automatic pipette should be used to obtain droplets of Matrigel for embedding in order to attenuate organoid-to-organoid morphological differences.
As a quality control, we recommend testing FOXA2/OTX2/LMX1A marker expression on day 20 to day 30 before performing the analysis on adult organoids, through RT-qPCR and immunofluorescence (Tables 1 and 2).
Troubleshooting
Please see Table 3 for a list of common problems with the protocols, their causes, and potential solutions.
Understanding Results
This protocol generates VM organoids that can be used to model both early and late phases of DA differentiation in a dish, from the formation of the mesencephalic floor plate to the generation of mature DA neurons capable of producing neuromelanin.
The morphological analysis of VM organoids at different developmental stages under a bright-field microscope is the first step in understanding the efficiency of differentiation. In fact, VM organoids change in size, shape, and cellular composition during differentiation. The formation of buds between days 8 and 12 in the outer organoid regions indicates that the floor plate is forming, and is a sign of correct differentiation (Fig. 2B and Fig. 3B-D). After morphological analysis, RT-qPCR is the most straightforward method to monitor the progress of VM organoid differentiation at a molecular level. We recommend analyzing the expression of early and late DA markers on days 15, 30, and 60 of differentiation to trace a reliable expression profile of VM organoid cultures.
FOXA2 , CORIN , EN1 , and SHH peak at day 30 and indicate the proper formation of DA neuron progenitors, which will go on to acquire mature molecular features and express markers such as NURR1 and TH at day 60 (Figs. 3A and 3F). Immunohistochemical analysis enables a more detailed evaluation of marker expression at the protein level and allows monitoring for the presence of specific cytoarchitecture within organoids. In fact, radially organized cell structures resembling rosettes are an important architectural element showing the correct formation of the mesencephalic floor plate (Fig. 3B-D). In line with RT-qPCR analysis, robust TH expression can only be detected at month 2 (Fig. 3E), indicating that cells are acquiring post-mitotic molecular characteristics. The detection of neuromelanin starting from month 3 using a bright-field microscope can further help characterize developing VM organoids and monitor differentiation efficiency. The release of neuromelanin is considered one of the last events in the maturation of human DA neurons. Morphological observations, combined with immunohistochemical analysis that can more precisely determine the release of neuromelanin, show that mature and pigmented DA neurons emerge within VM organoids (Fig. 4A-C).
Time Considerations
The preparation of hPSCs to start VM organoid differentiation (Basic Protocol 1) takes at least 10 days (including thawing, culturing, and one passage before beginning differentiation). The time required to generate VM organoids following Basic Protocol 2 can vary depending on the differentiation phase of interest. If the research question is focused on floor plate progenitor cells, the highest expression of FOXA2/OTX2/LMX1A occurs between days 20 and 30. If, instead, the research focus is on the maturation of DA neurons and cell-cell interaction studies with glial cells, 3 to 4 months are sufficient. For DA diversity studies or PD modeling using patient-derived iPSCs, at least 6 months are needed to obtain fully mature DA neurons with robust neuromelanin release. For VM organoid characterization at a molecular level (Basic Protocol 3), gene expression analysis to determine progenitor cell identity requires 1 to 2 days. Immunocytochemical analysis to determine progenitor cell identity requires 2 days.
Acknowledgments
The authors thank Ulla Jarl and Bengt Mattsson for their technical assistance. The research leading to these results was supported by funding from the New York Stem Cell Foundation, European Research Council (ERC) under ERC Grant Agreement 771427, European Union–funded project NSC-Reconstruct (European Union, H2O20, GA no 874758, 2020-23), Swedish Research Council (2016-00873), Swedish Parkinson Foundation (Parkinsonfonden), Swedish Brain Foundation, Strategic Research Area at Lund University Multipark, and Knut and Alice Wallenberg Stiftelse (KAW 2018-0040). Malin Parmar is a New York Stem Cell Foundation Robertson Investigator. Alessandro Fiorenzano is the recipient of grants from the Thorsten and Elsa Segerfalk Foundation, the Swedish Parkinson Foundation (Parkinsonfonden 1347/21) and the Anna-Lisa Rosenberg Foundation, which used to support this study.
Author Contributions
Edoardo Sozzi : Conceptualization, Data curation, Formal analysis, Investigation, Methodology, Visualization, Original draft; Fredrik Nilsson : Formal analysis, Investigation, Draft review and editing; Janko Kajtez : Visualization, Draft review and editing; Malin Parmar : Conceptualization, Funding acquisition, Resources, Supervision, Draft review and editing; Alessandro Fiorenzano : Conceptualization, Data curation, Funding acquisition, Methodology, Resources, Supervision, Original draft.
Conflict of Interest
MP is the owner of Parmar Cells AB and co-inventor of the following patents WO2016162747A2, WO2018206798A1, and WO2019016113A1. MP is a paid consultant, steering group member, and performs commissioned research for Novo Nordisk AS Cell Therapy Research and Development unit, and is a member of Arbor Bio SAB. The remaining authors declare no competing interests.
Open Research
Data Availability Statement
The data, tools, and materials (or their source) that support the protocol are available from the corresponding author upon reasonable request.
Literature Cited
- Agarwal, D., Sandor, C., Volpato, V., Caffrey, T. M., Monzon-Sandoval, J., Bowden, R., … Webber, C. (2020). A single-cell atlas of the human substantia nigra reveals cell-specific pathways associated with neurological disorders. Nature Communication , 11(1), 4183. doi: 10.1038/s41467-020-17876-0
- Arlotta, P., & Paşca, S. P. (2019). Cell diversity in the human cerebral cortex: From the embryo to brain organoids. Current Opinion in Neurobiology , 56, 194–198. doi: 10.1016/j.conb.2019.03.001
- Bjorklund, A., & Dunnett, S. B. (2007). Dopamine neuron systems in the brain: An update. Trends in Neuroscience (Tins) , 30(5), 194–202. doi: 10.1016/j.tins.2007.03.006
- Cederquist, G. Y., Asciolla, J. J., Tchieu, J., Walsh, R. M., Cornacchia, D., Resh, M. D., & Studer, L. (2019). Specification of positional identity in forebrain organoids. Nature Biotechnology , 37(4), 436–444. doi: 10.1038/s41587-019-0085-3
- Demers, C. J., Soundararajan, P., Chennampally, P., Cox, G. A., Briscoe, J., Collins, S. D., & Smith, R. L. (2016). Development-on-chip: In vitro neural tube patterning with a microfluidic device. Development , 143(11), 1884–1892. doi: 10.1242/dev.126847
- Fiorenzano, A., Birtele, M., Wahlestedt, J. N., & Parmar, M. (2021). Evaluation of TH-Cre knock-in cell lines for detection and specific targeting of stem cell-derived dopaminergic neurons. Heliyon , 7(1), e06006. doi: 10.1016/j.heliyon.2021.e06006
- Fiorenzano, A., Pascale, E., Gagliardi, M., Terreri, S., Papa, M., Andolfi, G., … Fico, A. (2018). An ultraconserved element containing lncRNA preserves transcriptional dynamics and maintains ESC self-renewal. Stem Cell Reports , 10(3), 1102–1114. doi: 10.1016/j.stemcr.2018.01.014
- Fiorenzano, A., Sozzi, E., Birtele, M., Kajtez, J., Giacomoni, J., Nilsson, F., … Parmar, M. (2021). Single-cell transcriptomics captures features of human midbrain development and dopamine neuron diversity in brain organoids. Nature Communication , 12(1), 7302. doi: 10.1038/s41467-021-27464-5
- Fiorenzano, A., Sozzi, E., Parmar, M., & Storm, P. (2021). Dopamine neuron diversity: Recent advances and current challenges in human stem cell models and single cell sequencing. Cells , 10(6), 1366. doi: 10.3390/cells10061366
- Jo, J., Xiao, Y. X., Sun, A. X., Cukuroglu, E., Tran, H. D., Goke, J., … Ng, H. H. (2016). Midbrain-like organoids from human pluripotent stem cells contain functional dopaminergic and neuromelanin-producing neurons. Cell Stem Cell , 19(2), 248–257. doi: 10.1016/j.stem.2016.07.005
- Kanton, S., Boyle, M. J., He, Z., Santel, M., Weigert, A., Sanchis-Calleja, F., … Camp, J. G. (2019). Organoid single-cell genomic atlas uncovers human-specific features of brain development. Nature , 574(7778), 418–422. doi: 10.1038/s41586-019-1654-9
- Kelava, I., & Lancaster, M. A. (2016). Stem cell models of human brain development. Cell Stem Cell , 18(6), 736–748. doi: 10.1016/j.stem.2016.05.022
- Kim, J., Koo, B. K., & Knoblich, J. A. (2020). Human organoids: Model systems for human biology and medicine. Nature Reviews Molecular Cell Biology , 21(10), 571–584. doi: 10.1038/s41580-020-0259-3
- Krishnaswami, S. R., Grindberg, R. V., Novotny, M., Venepally, P., Lacar, B., Bhutani, K., … Lasken, R. S. (2016). Using single nuclei for RNA-seq to capture the transcriptome of postmortem neurons. Nature Protocols , 11(3), 499–524. doi: 10.1038/nprot.2016.015
- Lancaster, M. A., Renner, M., Martin, C. A., Wenzel, D., Bicknell, L. S., Hurles, M. E., … Knoblich, J. A. (2013). Cerebral organoids model human brain development and microcephaly. Nature , 501(7467), 373–379. doi: 10.1038/nature12517
- Miura, Y., Li, M. Y., Birey, F., Ikeda, K., Revah, O., Thete, M. V., … Pasca, S. P. (2020). Generation of human striatal organoids and cortico-striatal assembloids from human pluripotent stem cells. Nature Biotechnology , 38(12), 1421–1430. doi: 10.1038/s41587-020-00763-w
- Nilsson, F., Storm, P., Sozzi, E., Hidalgo Gil, D., Birtele, M., Sharma, Y., … Fiorenzano, A. (2021). Single-cell profiling of coding and noncoding genes in human dopamine neuron differentiation. Cells , 10(1), 137. doi: 10.3390/cells10010137
- Nolbrant, S., Heuer, A., Parmar, M., & Kirkeby, A. (2017). Generation of high-purity human ventral midbrain dopaminergic progenitors for in vitro maturation and intracerebral transplantation. Nature Protocols , 12(9), 1962–1979. doi: 10.1038/nprot.2017.078
- Pasca, S. P. (2018). The rise of three-dimensional human brain cultures. Nature , 553(7689), 437–445. doi: 10.1038/nature25032
- Quadrato, G., Nguyen, T., Macosko, E. Z., Sherwood, J. L., Min Yang, S., Berger, D. R., … Arlotta, P. (2017). Cell diversity and network dynamics in photosensitive human brain organoids. Nature , 545(7652), 48–53. doi: 10.1038/nature22047
- Rifes, P., Isaksson, M., Rathore, G. S., Aldrin-Kirk, P., Moller, O. K., Barzaghi, G., … Kirkeby, A. (2020). Publisher correction: Modeling neural tube development by differentiation of human embryonic stem cells in a microfluidic WNT gradient. Nature Biotechnology , 38(11), 1357. doi: 10.1038/s41587-020-0590-4
- Siddiqi, S. H., Kording, K. P., Parvizi, J., & Fox, M. D. (2022). Causal mapping of human brain function. Nature Reviews Neuroscience , 23(6), 361–375. doi: 10.1038/s41583-022-00583-8
- Tiklova, K., Nolbrant, S., Fiorenzano, A., Bjorklund, A. K., Sharma, Y., Heuer, A., … Parmar, M. (2020). Single cell transcriptomics identifies stem cell-derived graft composition in a model of Parkinson's disease. Nature Communications , 11(1), 2434. doi: 10.1038/s41467-020-17421-z
Corrections
In this publication, the following correction has been made:
In step 14 of Basic Protocol 2, the concentration of SB431542 has been changed to 10 µM.
The current version online now includes this correction and may be considered the authoritative version of record.
Citing Literature
Number of times cited according to CrossRef: 11
- Jessica Cohen, Annette Mathew, Kirk D. Dourvetakis, Estella Sanchez-Guerrero, Rajendra P. Pangeni, Narasimman Gurusamy, Kristina K. Aenlle, Geeta Ravindran, Assma Twahir, Dylan Isler, Sara Rukmini Sosa-Garcia, Axel Llizo, Alison C. Bested, Theoharis C. Theoharides, Nancy G. Klimas, Duraisamy Kempuraj, Recent Research Trends in Neuroinflammatory and Neurodegenerative Disorders, Cells, 10.3390/cells13060511, 13 , 6, (511), (2024).
- Valentina Basso, Máté D. Döbrössy, Lachlan H. Thompson, Deniz Kirik, Heidi R. Fuller, Monte A. Gates, State of the Art in Sub-Phenotyping Midbrain Dopamine Neurons, Biology, 10.3390/biology13090690, 13 , 9, (690), (2024).
- Harysh Winster Suresh Babu, Sindduja Muthu Kumar, Harsimrat Kaur, Mahalaxmi Iyer, Balachandar Vellingiri, Midbrain organoids for Parkinson's disease (PD) - A powerful tool to understand the disease pathogenesis, Life Sciences, 10.1016/j.lfs.2024.122610, 345 , (122610), (2024).
- Yuan-Wei Yan, Eddie S Qian, Lauren E Woodard, Julie Bejoy, Neural lineage differentiation of human pluripotent stem cells: Advances in disease modeling, World Journal of Stem Cells, 10.4252/wjsc.v15.i6.530, 15 , 6, (530-547), (2023).
- Naoto Sugeno, Takafumi Hasegawa, Unraveling the Complex Interplay between Alpha-Synuclein and Epigenetic Modification, International Journal of Molecular Sciences, 10.3390/ijms24076645, 24 , 7, (6645), (2023).
- Yee Jie Yeap, Tng J. W. Teddy, Mok Jung Lee, Micaela Goh, Kah Leong Lim, From 2D to 3D: Development of Monolayer Dopaminergic Neuronal and Midbrain Organoid Cultures for Parkinson’s Disease Modeling and Regenerative Therapy, International Journal of Molecular Sciences, 10.3390/ijms24032523, 24 , 3, (2523), (2023).
- Hilary S Y Toh, Xin Yi Choo, Alfred Xuyang Sun, Midbrain organoids—development and applications in Parkinson’s disease, Oxford Open Neuroscience, 10.1093/oons/kvad009, 2 , (2023).
- Babak Rezaei, Jessica Giacomoni, Fredrik Nilsson, Edoardo Sozzi, Alessandro Fiorenzano, Malin Parmar, Stephan S Keller, Janko Kajtez, Modular 3D printed platform for fluidically connected human brain organoid culture, Biofabrication, 10.1088/1758-5090/ad0c2c, 16 , 1, (015014), (2023).
- Denis Pavăl, The dopamine hypothesis of autism spectrum disorder: A comprehensive analysis of the evidence, Fragile X and Related Autism Spectrum Disorders, 10.1016/bs.irn.2023.08.009, (1-42), (2023).
- Anna Kreutz, Guang Hu, Erik Tokar, Pluripotent Stem Cell-derived Dopaminergic Neurons for Studying Developmental Neurotoxicity, Stem Cell Reviews and Reports, 10.1007/s12015-023-10555-9, 19 , 7, (2120-2130), (2023).
- Edoardo Sozzi, Janko Kajtez, Andreas Bruzelius, Milan Finn Wesseler, Fredrik Nilsson, Marcella Birtele, Niels B. Larsen, Daniella Rylander Ottosson, Petter Storm, Malin Parmar, Alessandro Fiorenzano, Silk scaffolding drives self-assembly of functional and mature human brain organoids, Frontiers in Cell and Developmental Biology, 10.3389/fcell.2022.1023279, 10 , (2022).

