Fixation and imaging of HeLa cells after mitochondrial depolarization
OLIVIA HARDING, Erika L.F. Holzbaur
Abstract
Ectopic expression of p62/SQSTM1 often induces puncta formation and poor cell health due to p62’s proclivity to multimerize and form filaments. We used the cell fixation protocol described here in order to over-express fluorescently tagged p62 at low levels in p62-/- cells and amplify the signal with immuno-labeling. By this method, we were able to express a variety of tagged p62 mutations to determine their effects on NEMO recruitment to depolarized mitochondria without introducing toxic artifacts of p62 overexpression. We also note the use of a modified fixation protocol to visualize various endogenous ATG8 proteins, which are also difficult to over-express in our system. Thus, fixation techniques are a critical complement to studies in live cells.
Before start
-
The start point for this protocol is after cells grown on glass coverslips in a 12-well plate have been transfected with Parkin, EGFP-NEMO, HaloOPTN, and mCherry-p62 for 18-
24h 0m 0sand tagged with Halo ligand. -
Prepare
45millimolar (mM)stock of Antimycin A by suspending50mgsolid AntA in2mLethanol. -
Prepare
10millimolar (mM)stock Oligomycin A by suspending5mgsolid OligA in630µLDMSO -
Prepare 4% PFA in PBS. Keep frozen at
-20°C.NotePrepare fresh for day-of fixation or thaw directly before use.Will use 1mL 4% PFA per well. -
Prepare Culture Media by making 10% FBS, 1% GlutaMAX solution in DMEM.
-
Prepare 0.5% Triton X-100 in PBS (Permeabilization buffer).
NotePrepare 0.75mL Permeabilization buffer per well, store at 4°C.Bring to Room temperature before use. This will provide less shock to cells, better preserving fixed structures.Do not use Triton for permeabilization if LC3- autophagosomes are the structure of interest, since Triton is too harsh. Use ice cold methanol if imaging LC3 structures -
Prepare 0.2% Triton X-100/3% BSA in PBS (Blocking buffer).
NotePrepare 0.75mL Blocking buffer per well, store at 4°C.Bring to Room temperature before use. -
Prepare a humidity chamber as previously described (dx.doi.org/10.17504/protocols.io.bujsnune)
Attachments
Steps
AntA/OligA treatment
Prepare working AntA/OligA solution by transferring 0.5mL conditioned media to a 1.5 mL tube and adding 0.25µL 10millimolar (mM) OligA and 2mL 45millimolar (mM) AntA.
Gently drop working AntA/OligA solution onto cells.
Incubate at 37°C, 5% CO2 for 0h 55m 0s- 1h 5m 0s.
Fixation
25 minutes before AntA/OligA treatment is complete, warm 4% PFA and 1X PBS to 37°C.
Remove cells from incubator and aspirate media.
Quickly drop on 0.5mL warmed 1X PBS.
Aspirate PBS.
Repeat warm PBS wash.
Drop on 1mL warmed 4% PFA.
Incubate at 37°C for 0h 10m 0s.
Drop on 0.5mL warmed PBS.
Aspirate PBS.
Drop on 0.5mL warmed PBS.
Incubate covered at Room temperature for 0h 3m 0s.
Permeabilization
Aspirate PBS.
Drop on 0.75mL Permeabilization buffer.
Incubate covered at Room temperature for 0h 5m 0s.
Blocking
Add ~250µL Blocking buffer to each well.
Incubate covered at Room temperature for 0h 45m 0s.
Aspirate Blocking buffer.
Primary antibody
Prepare 200µL Primary antibody dilution per coverslip, with 1:250 antiHSP60 and 1:500 anti-p62 in Blocking buffer.
Use sharp forceps to carefully lift the coverslip out of the well and dab excess Blocking buffer on a Kimwipe.
Place slip cell-side up on Parafilm in the humidity chamber.
From the edge of the coverslip, pipet primary antibody dilution onto the cells.
Incubate covered at 4°C 0h 5m 0s.
18- 24 hours later, aspirate antibody dilution from the edge of the coverslip, gently lifting one side of the slip with forceps if necessary to allow buffer to slide off.
From the edge of the coverslip, pipet 100µL Room temperature PBS onto the cells.
Incubate covered at Room temperature for 0h 5m 0s.
Aspirate PBS from the edge of the coverslip, gently lifting one side of the slip with forceps if necessary to allow buffer to slide off.
Repeat previous three steps for a total of four washes.
Wash (1/4):
- From the edge of the coverslip, pipet
100µLRoom temperaturePBS onto the cells. - Incubate covered at
Room temperaturefor0h 5m 0s. - Aspirate PBS from the edge of the coverslip, gently lifting one side of the slip with forceps if necessary to allow buffer to slide off.
Wash (2/4):
- From the edge of the coverslip, pipet
100µLRoom temperaturePBS onto the cells. - Incubate covered at
Room temperaturefor0h 5m 0s. - Aspirate PBS from the edge of the coverslip, gently lifting one side of the slip with forceps if necessary to allow buffer to slide off.
Wash (3/4):
- From the edge of the coverslip, pipet
100µLRoom temperaturePBS onto the cells. - Incubate covered at
Room temperaturefor0h 5m 0s. - Aspirate PBS from the edge of the coverslip, gently lifting one side of the slip with forceps if necessary to allow buffer to slide off.
Wash (4/4):
- From the edge of the coverslip, pipet
100µLRoom temperaturePBS onto the cells. - Incubate covered at
Room temperaturefor0h 5m 0s. - Aspirate PBS from the edge of the coverslip, gently lifting one side of the slip with forceps if necessary to allow buffer to slide off.
Secondary antibody
Prepare secondary antibody solution by diluting goat anti-rabbit 405 and goat anti-mouse 1:200 in Blocking buffer.
From the edge of the coverslip, pipet secondary antibody dilution onto the cells.
Incubate covered at Room temperature for 0h 45m 0s.
Aspirate antibody dilution from the edge of the coverslip, gently lifting one side of the slip with forceps if necessary to allow buffer to slide off.
From the edge of the coverslip, pipet 100µL Room temperature PBS onto the cells.
Incubate covered at Room temperature for 0h 5m 0s.
Aspirate PBS from the edge of the coverslip, gently lifting one side of the slip with forceps if necessary to allow buffer to slide off.
Repeat previous three steps for a total of four washes.
Wash (1/4)
- From the edge of the coverslip, pipet
100µLRoom temperaturePBS onto the cells. - Incubate covered at
Room temperaturefor0h 5m 0s. - Aspirate PBS from the edge of the coverslip, gently lifting one side of the slip with forceps if necessary to allow buffer to slide off.
Wash (2/4)
- From the edge of the coverslip, pipet
100µLRoom temperaturePBS onto the cells. - Incubate covered at
Room temperaturefor0h 5m 0s. - Aspirate PBS from the edge of the coverslip, gently lifting one side of the slip with forceps if necessary to allow buffer to slide off.
Wash (3/4)
- From the edge of the coverslip, pipet
100µLRoom temperaturePBS onto the cells. - Incubate covered at
Room temperaturefor0h 5m 0s. - Aspirate PBS from the edge of the coverslip, gently lifting one side of the slip with forceps if necessary to allow buffer to slide off.
Wash (4/4)
- From the edge of the coverslip, pipet
100µLRoom temperaturePBS onto the cells. - Incubate covered at
Room temperaturefor0h 5m 0s. - Aspirate PBS from the edge of the coverslip, gently lifting one side of the slip with forceps if necessary to allow buffer to slide off.
Mounting
Pipet 12.5µL Room temperature VectaShield onto a microscope slide.
Pick up coverslip with forceps and dab excess PBS onto a Kimwipe.
Lay coverslip cell-side down onto VectaShield drop.
Seal edges of coverslip with nail polish.
Lay flat until set.
Imaging
Using 60X objective and RFP epifluorescence, find the focal plane of fixed cells by looking for dsRed2-labeled mitochondria.
Configure 405, 488, 561, and 647 lasers and accompanying exposure times to maximize dynamic range of each signal.
Configure collection parameters to record a >2 um Z stack of images.
Collect images from fields of view with multiple cells exhibiting all visible tags. Collect volume from the 50th to 75th percentile of the cell (see Figure 1). If mitochondria are depolarized, can use OPTN signal (recruitment to
fragmented mitochondria) to determine whether Parkin is expressed.
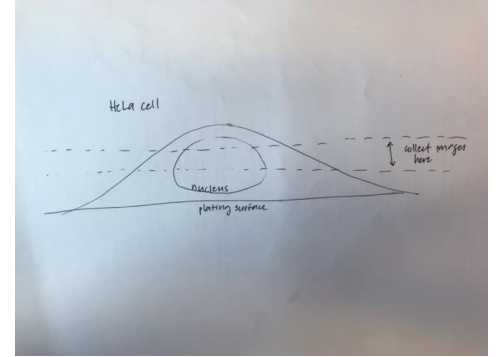
Collect images in order to record 10-20 cells per condition, depending on the quality of the transfection and cells. Collect images from diverse areas of the coverslip.

