Extraction of Total Nucleic Acid from Wastewater Using the Promega Wizard Enviro Total Nucleic Acid Kit
Padmini Ramachandran, Tamara Walsky, Amanda Windsor, Maria Hoffmann, Christopher Grim
Abstract
Wastewater based epidemiology has proven to be a useful tool in the COVID-19 pandemic, allowing prevalence and variant and sub-lineage surveillance on a watershed population level in a non-intrusive manner. Essential to this method is the efficient extraction of viral genomic material from wastewater. The process of detecting SARS Co-V-2 genetic signatures in wastewater samples involves collection of water, either as a grab sample or as a 24-hour composite sample, followed by sample concentration. Target genetic material may be present at a low concentration in water samples, making sample concentration a prerequisite for sensitive detection. Concentration of microbial matter can be performed using a variety of methods, such as charged membrane filtration, centrifugal ultrafiltration and flocculation/precipitation using skim milk or polyethylene glycol (PEG)/NaCl. Most of the concentration methods were originally developed to concentrate live matter with the objective of culturing for detection of intact particles, though they have also been used for PCR-based detection. These methods have proven to be inconsistent, labor intensive and time consuming.
More and more SC2-wastewater specific methods have recently been developed. We have assessed the Promega Wizard Enviro Total Nucleic Acid kit for extraction of RNA for downstream SC2 detection and sequencing. Briefly, the method directly captures and concentrates total nucleic acids (TNA) from a large volume of water using PureYield™ columns. The method uses a short protocol that minimizes the need for specialized laboratory equipment. In a first step total nucleic acid from a large volume sample (e.g., 40ml of wastewater) is captured on a PureYield™ Binding Column and then eluted in 1ml. In a second step, the material is further purified and concentrated using the PureYield™ Minicolumn. This method achieves consistent recovery rates and significant reduction in PCR inhibitors.
The total nucleic acid extracted using this kit can be analyzed for SARS-CoV-2 targets using a SARS-CoV-2 RT-qPCR
kit for wastewater. Please visit the Promega website for more information on these products:
https://www.promega.com/applications/infectious-diseases/covid19-wastewater-sars-cov-2-detection/. Additionally, the extracted RNA can be utilized for SC2 variant sequencing: Enhanced QIAseq DIRECT SARS-CoV-2 Kit for Illumina MiSeq (protocols.io), or Modified NEBNext® VarSkip Short SARS-CoV-2 Library Prep Kit for Illumina Platforms - adapted for wastewater samples (protocols.io).
Before start
Prepare the following solutions prior to beginning nucleic acid extraction in Section 4:
Column Wash 1 (CWE): Add 57mLof isopropanol to the Column Wash 1 (CWE) bottle and mark on the bottle “plus
isopropanol”. This reagent is stable at 15°C to 30°C when tightly capped.
Column Wash 2 (RWA): Add 350mLof 95% (v/v) ethanol the Column Wash 2 (RWA) bottle and mark the bottle “plus ethanol”. This reagent is stable at 15°C to 30°C when tightly capped.
Turn on water bath to 60°C
Steps
Capture and Concentration
Dispense 40mL of wastewater or pasteurized wastewater into a
Equipment
| Value | Label |
|---|---|
| 50 mL conical screw-cap tube | NAME |
| Plastic consumables | TYPE |
| Grenier Bio-One | BRAND |
| 5622-7261 | SKU |
| Any brand or size of plastic tube with a tight-fitting screw cap will work fine. | SPECIFICATIONS |
Add 500µL of Protease Solution. Mix well by inversion and incubate for 0h 30m 0s at Room temperature .
3000x g,25Room temperature .
Carefully decant 20mLof the supernatant into each of two clean
Equipment
| Value | Label |
|---|---|
| 50 mL conical screw-cap tube | NAME |
| Plastic consumables | TYPE |
| Grenier Bio-One | BRAND |
| 5622-7261 | SKU |
| Any brand or size of plastic tube with a tight-fitting screw cap will work fine. | SPECIFICATIONS |
Discard the 50ml conical tube containing the pellet into an appropriate biohazard waste container.
To each tube containing 20mLof the clarified supernatant, add 6mLof Binding Buffer 1 (BBD) followed by
500µLof Binding Buffer 2 (BBE).
Mix well by inverting the tube gently 10 times, or until thoroughly mixed.
Add 24mLof isopropanol to each tube.
Mix well by inverting the tube gently 10 times, or until thoroughly mixed.
Setup the vacuum manifold as follows (see Figure 1): remove vacuum port cap, attach a Reservoir Extension Funnel to the PureYield™ Binding Column, then connect the column to the vacuum manifold by pressing the nozzle gently into the vacuum port.
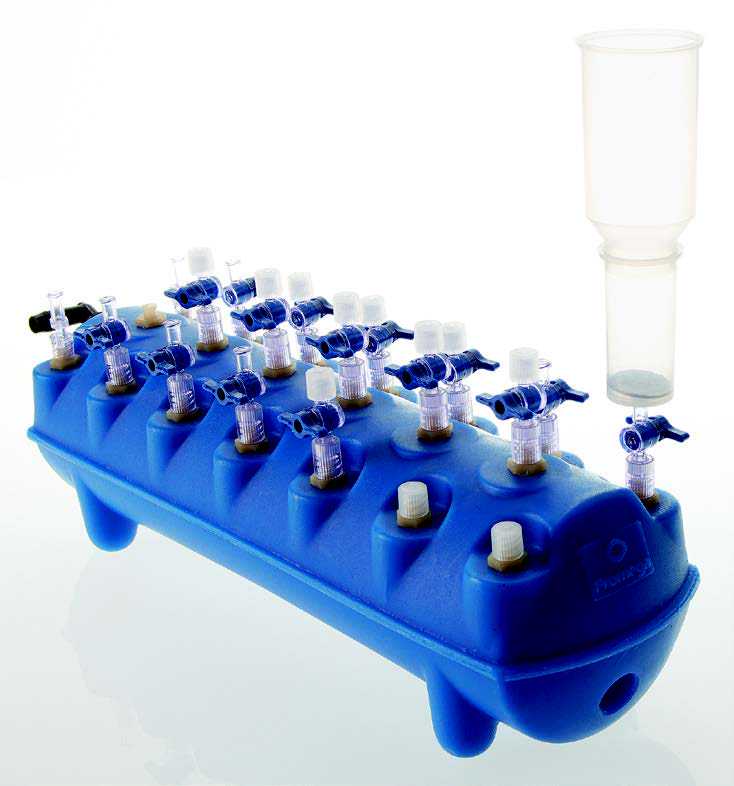
Pour the mixture from each tube from Step 8 into the Reservoir Extension Funnel on the PureYield™ Binding
Column (combine both tubes of the same sample if applicable), turn on the pump and apply vacuum to capture
TNA on the column.
Add 5mLof Column Wash 1 (CWE) and apply a vacuum to pull the liquid through the PureYield™ Binding
Column.
Add 20mLof Column Wash 2 (RWA) and apply a vacuum to pull the liquid through the PureYield™ Binding
Column. Continue to draw a vacuum for an additional 0h 0m 30s after all visible liquid has passed through the
membrane.
Release the vacuum and remove the column from the vacuum manifold. Preheat 1.2mLof Nuclease-Free Water,
per sample, to 60°C for 0h 5m 0s.
Assemble the elution device by placing a 1.5ml microcentrifuge tube into the base of the Eluator™ Vacuum
Elution Device (Cat.# A1071) and securing the tube cap in the open position, as shown (Figure 2).
Insert the PureYield™ Binding Column into the top of the Eluator Device, making sure the column is fully seated
on the collar as shown in Figure 3.


Place the Eluator™ Device assembly onto a vacuum manifold (Figure 3). Add 500µLof preheated (60°C)
Nuclease-Free Water to the PureYield™ Binding Column. Apply maximum vacuum for 0h 1m 0sor until all
liquid has passed through the column. Repeat the process by adding another 500µLof preheated Nuclease-Free
Water to the PureYield™ Binding Column to elute a total of 1mLof TNA solution.
TNA Extraction and Clean-Up
Add 400µLof Binding Buffer 1(BBD) and 100µLof Binding Buffer 2 (BBE) to 1mLof liquid eluted in
Step 15.
Mix well by inverting the tube gently 10 times, or until thoroughly mixed. Divide the contents into two
Equipment
| Value | Label |
|---|---|
| DNA LoBind Tube 1.5 mL | NAME |
| Microcentrifuge tube | TYPE |
| Eppendorf | BRAND |
| 022431021 | SKU |
containing 750µLeach.
0h 1m 0s Add 750µLof isopropanol to each tube and mix well by inverting the tube gently 10 times, or until thoroughly mixed.
Place the PureYield™ Minicolumn into a PureYield™ Collection Tube. Pass the entire volume of the mixture
through the column, 750µLat a time (a total of four times), using a microcentrifuge at 10.000rpm.
Add 300µLof Column Wash 1 (CWE) and pull through the PureYield™ Minicolumn by centrifugation 10000rpm . Discard the flow-through.
Add 500µLof Column Wash 2 (RWA) and pull through the PureYield™ Minicolumn by centrifugation 10000rpm . Repeat this wash one time. Discard the flowthrough.
Centrifuge 10000rpm to remove any residual wash solution.
Preheat 65µLof Nuclease-Free Water per sample to 60°C for 0h 5m 0s.
Transfer the PureYield™ Minicolumn to a new
Equipment
| Value | Label |
|---|---|
| DNA LoBind tubes, 1.5 mL | NAME |
| Tubes | TYPE |
| Eppendorf | BRAND |
| 022431021 | SKU |
| 1.5 mL | SPECIFICATIONS |
and add 30µLof preheated (60°C) Nuclease-Free Water to the column. Let the water soak into the column filter for approximately 0h 1m 0s.
Centrifuge 10000rpm to elute. Repeat elution with another 30µLof preheated Nuclease-Free Water, for a total of 60µL.
Store sample at or below -20°C until further analysis. TNA purified using this method can be directly used for
RT-qPCR, for example as input into the following protocol:

