Expression and Purification of Human U1-70K (snRNP70) and its BAD Domains Using an E. coli Expression System
Zihan Zhang, Zihan Zhang, Jun Zhang, Jun Zhang, Trenton M. Paul, Trenton M. Paul, Shariq Jamal, Shariq Jamal, Ethan Ekpenyong, Ethan Ekpenyong, Peter Prevelige, Peter Prevelige, Talia E. Fargason, Talia E. Fargason
Abstract
U1-70K (snRNP70) serves as an indispensable protein component within the U1 complex, assuming a pivotal role in both constitutive and alternative RNA splicing processes. Notably, U1-70K engages in interactions with SR proteins, instigating the assembly of the spliceosome. This protein undergoes regulation through phosphorylation at multiple sites. Of significant interest, U1-70K has been implicated in Alzheimer's disease, in which it tends to form detergent-insoluble aggregates. Even though it was identified more than three decades ago, our understanding of U1-70K remains notably constrained, primarily due to challenges such as low levels of recombinant expression, susceptibility to protein degradation, and insolubility. In endeavoring to address these limitations, we devised a multifaceted approach encompassing codon optimization, strategic purification, and a solubilization protocol. This methodology has enabled us to achieve a high yield of full-length, soluble U1-70K, paving the way for its comprehensive biophysical and biochemical characterization. Furthermore, we provide a detailed protocol for the preparation of phosphorylated U1-70K. This set of protocols promises to be a valuable resource for scientists exploring the intricate web of U1-70K-related mechanisms in the context of RNA splicing and its implications in neurodegenerative disorders and other disorders and biological processes. © 2024 The Authors. Current Protocols published by Wiley Periodicals LLC.
Basic Protocol 1 : Expression and purification of full-length U1-70K from E. coli
Support Protocol 1 : Making chemically competent BL21 Star pRARE/pBB535 cells
Basic Protocol 2 : Phosphorylation of full-length U1-70K using SRPK1
Support Protocol 2 : Purification of SRPK1
Basic Protocol 3 : Expression and purification of U1-70K BAD1 from E. coli
Basic Protocol 4 : Phosphorylation of U1-70K BAD1 using SRPK1
Basic Protocol 5 : Expression and purification of U1-70K BAD2 from E. coli
INTRODUCTION
Most eukaryotic RNA transcripts undergo splicing on the spliceosome, which is a dynamic protein-RNA complex composed of U1, U2, U4, U5, and U6 snRNPs. Spliceosome assembly starts with recruitment of U1 snRNP to the 5′ splicing site on the intron. U1-70K (also known as snRNP70) is the pivotal component of the U1 snRNP complex involved in initiating spliceosome assembly by interacting with SR proteins bound to the 5′ splicing site (Moore, 1993).
U1-70K consists of a well-structured N-terminal RNA recognition motif (RRM) and two C-terminal basic acidic dipeptide repeat domains (BAD1 and BAD2). The U1-70K RRM specifically interacts with the U1 snRNA and integrates the protein into the U1 snRNP (Kondo et al., 2015). The functions of BAD domains are unclear, although recent studies have implicated BAD domains in U1-70K's phase separation and aberrant aggregation in Alzheimer's disease (Bishof et al., 2018; Diner et al., 2014; Xue et al., 2019). In addition, BAD domains possess extensive serine residues, consistent with the facts that U1-70K is heavily phosphorylated in cells and that blocking dephosphorylation of U1-70K abolishes RNA splicing (Tazi et al., 1993; Woppmann et al., 1990).
In addition to its functions in RNA splicing, U1-70K is also involved in RNA nuclear retention, degradation, and polyadenylation (Hu et al., 2022; Lee et al., 2022). A recent cryo-EM study revealed that U1-70K interacts directly with RNA polymerase II, suggestive of a role in RNA transcription (Zhang et al., 2020). However, the flexible nature of U1-70K's BAD1 and BAD2 domains renders them undetectable by cryo-EM, leaving their roles in these interactions elusive (Zhang et al., 2020). U1-70K is also implicated in neurodegenerative and autoimmune diseases (Diner et al., 2014; Kattah et al., 2010).
Despite its involvement in RNA splicing and various diseases, our understanding of U1-70K is still limited. The literature also presents a discrepancy in regard to whether U1-70K's recruitment to pre-mRNA is mediated by its RRM or its BAD1 domains (Cao & Garcia-Blanco, 1998; Cho et al., 2011). Comprehensive understanding of U1-70K functions and clarification of this controversy will require in-depth biophysical and biochemical characterization. However, this has been hindered by challenges in overexpression and purification of the full-length protein for decades. Earlier attempts to express U1-70K in Escherichia coli have shown this to be challenging (Northemann et al., 1995). Co-expression with the chaperones DnaK and DnaJ has facilitated detectable levels of full-length U1-70K expression (Richter et al., 2014). Despite these efforts, however, successful purification of active, full-length U1-70K or its individual BAD1 or BAD2 domains has remained unachieved.
Here, we introduce a protocol for the overexpression and purification of full-length U1-70K, and its BAD1 and BAD2 domains, in E. coli. Subsequent in vitro phosphorylation of these proteins is also detailed. The ability to purify active U1-70K and its BAD domains, both phosphorylated and unphosphorylated, from E. coli paves the way for comprehensive in vitro studies of these molecules. These include investigating U1-70K's interactions with RNA and other proteins and elucidating the roles of the BAD domains and phosphorylation in these interactions. This article contains five basic protocols for five processes: first, the purification of full-length wild type U1-70K using an E.coli expression system; second, the phosphorylation of the full-length wild-type U1-70K using SRPK1 in vitro ; third, the purification of the U1-70K BAD1 domain using and E. coli expression system; fourth, phosphorylation of the U1-70K BAD1 domain in vitro using SRPK1; and fifth, purification of the U1-70K BAD2 domain using an E. coli expression system. Purifications are accomplished through the use of chaperones to optimize expression followed by nickel and SP columns to purify the protein. Proteins are phosphorylated by incubation with SRPK1 for 1 hr for the BAD1 domain and 24 hr for the full-length protein.
Basic Protocol 1: EXPRESSION AND PURIFICATION OF FULL-LENGTH U1-70K FROM E. coli
This protocol details the expression of full-length human U1-70K (comprising residues 1-437) with an N-terminal SUMO tag using BL21 Star(DE3) cells coexpressing DnaK and DnaJ (encoded by plasmid pBB535). Coexpression of DnaK and DnaJ increases the yield of full-length U1-70K. In addition, the pRARE plasmid that encodes rare codons in the commercial Rosetta transformation-competent cell line is also needed to boost the yield of U1-70K. Therefore, a competent cell strain hosting both pBB535 and pRARE should be prepared first (Support Protocol 1). We then detail a technique for purifying the protein using Ni affinity chromatography and SP cation-exchange chromatography. The typical yield of U1-70K is ∼0.4 mg per liter of culture.
Materials
-
BL21 Star pRARE/pBB535 cells (chemically competent; see Support Protocol 1)
-
pSMT3 plasmid encoding human U1-70K (codon-optimized; available from the authors upon request)
-
Lysogeny broth (LB) medium (see recipe)
-
LB agar, Miller (Fisher cat. no. BP1425-2)
-
Kanamycin (Fisher cat. no. 50-213-386) or kanamycin stock (see recipe)
-
Chloramphenicol (ACROS cat. no. AC227920250) or chloramphenicol stock (see recipe)
-
Spectinomycin (Fisher cat. no. BP29571) or spectinomycin stock (see recipe)
-
Terrific broth (TB) medium (see recipe)
-
Deionized water or Milli-Q water
-
Lysis buffer (see recipe)
-
Low-salt wash 1 (see recipe)
-
Elution buffer 1 (see recipe)
-
SPA 1 buffer (see recipe)
-
SPB 1 buffer (see recipe)
-
Storage buffer (see recipe)
-
IPTG (Fisher cat. no. BP1755-100)
-
4× protein loading buffer (Gallagher, 2007)
-
HisPur Ni-NTA resin (Thermo Scientific cat. no. 88222)
-
2-, 10-, 20-, 200-, and 1000-µl pipets
-
10-, 200-, and 1000-µl pipet tips
-
Water bath (Fisher CPD02 or equivalent)
-
Incubator (VWR)
-
Petri dish (Fisher cat. no. FB087579B)
-
Plate spreader (Fisher cat. no. 14-665-230)
-
2.8-L and 250-ml culture flasks
-
Incubator shaker (New Brunswick Excella E25 or equivalent)
-
2-, 10-, and 25-ml serological pipets
-
Inoculating loops or needles (Fisher cat. no. 22-363-595)
-
Centrifuge (Avanti JXN-26 or equivalent)
-
Centrifuge bottles
-
50-ml conical centrifugal tube (Falcon cat. no. 14-959-49A)
-
15-ml conical centrifugal tube (Thermo Scientifics cat. no. 339650)
-
PVDF membrane filter, 0.22 μm pore size (Millipore cat. no. GVWP04700)
-
NanoDrop (One C) or equivalent spectrophotometer
-
Centrifuge with Beckman JA-8000 rotor
-
−80°C freezer
-
Sonicator (QSONICA Q500)
-
ÄKTA Start protein purification system (Cytiva) or equivalent
-
5-ml HiTrap SP HP column (Cytiva cat. no. 17115101)
-
10-kDa spin concentrators (Millipore Sigma cat. no. UFC901096)
-
1.5-ml low-protein-binding microcentrifuge tubes (Thermo Fisher Scientific cat. no. 90410)
-
Additional reagents and equipment for SDS-PAGE (Gallagher, 2007)
Day 1
Transform competent cells and plate them to select transformed cells
1.Thaw chemically competent BL21 Star pRARE/pBB535 cells on ice.
2.Add 1 μl pSMT3-U1-70K plasmid to the cells and incubate for 15 min on ice.
3.Heat shock the cells for 60 s at 42°C using a water bath.
4.Add 150 μl LB to the cells and allow to shake for 1 hr at 37°C.
5.Transfer cells to an agar plate with 50 μg/ml kanamycin, 50 μg/ml chloramphenicol, and 50 μg/ml spectinomycin. Spread until dry.
6.Incubate plate overnight at 37°C.
Prepare medium
7.Prepare 6.05 L of TB.
8.Add 1 L TB to 6 clean 2.8-L culture flasks, and add 50 ml TB to one 250-ml culture flask.
9.Autoclave TB.
10.Add antibiotics (50 μg/ml kanamycin, 50 μg/ml chloramphenicol, and 50 μg/ml spectinomycin) to the 50 ml of TB (starter culture).
Day 2
Prepare starter culture
11.Pick colonies from the agar plate with inoculating loops and use to inoculate the 50-ml starter culture.
12.Incubate starter culture overnight at 37°C with shaking at 220 RPM.
Prepare buffers
13.Prepare and filter all buffers: lysis buffer, low-salt wash 1, elution buffer 1, SPA 1, SPB 1, and storage buffer.
14.Filter all buffers through 0.22-μm filter membranes to prepare them for use in the ÄKTA system.
Day 3
Grow and induce the cells
15.Check the optical density of the starter culture at a wavelength of 600 nm (OD600).
16.Calculate the volume of starter culture needed to achieve a starting OD600 of 0.02 in 1 L TB.
17.Add antibiotics (50 μg/ml kanamycin, 50 μg/ml chloramphenicol, and 50 μg/ml spectinomycin) to each 1-L TB cultures.
18.Inoculate the TB with the calculated volume of starter culture from step 16.
19.Incubate at 37°C, 220 RPM, until the culture reaches an OD600 of 0.8-1.0.
20.Add 500 μl of 1 M IPTG to each flask.
21.Reduce the temperature to 25°C and continue incubating the cultures with 220 RPM shaking overnight (∼16 hr).
Day 4
Harvest the cells
22.Collect cells using a Beckman JA-8000 rotor for 15 min at 3000 RCF, 4°C.
23.Decant the supernatant and use a rubber spatula to collect cell pellets into a 50-ml Falcon tube.
24.Properly dispose of biohazardous medium. Be sure to consult your department or institution prior to conducting the protocol.
25.Check the mass of cells collected.
26.Freeze the cell pellet.
27.Thaw the cells.
Purify the protein
28.Resuspend cells with 1.5 ml lysis buffer per gram of cells.
29.Freeze/thaw cells twice: Incubate the sample in a − 80°C freezer for 20 min to freeze, then incubate the sample in a room-temperature water bath until cells have thawed (∼5 min), and repeat the procedure.
30.Thaw the cells.
31.Sonicate cells in a metal (or plastic) beaker using an ice-water bath to keep cool: Set the sonicator (1/2-inch tip) to 70% power, pulsing for 3 s on and 5 off, and sonicate for three to six cycles each comprising a total of 1 min of sonication, allowing the sample to cool for 5 min between rounds.
32.Spin down the cell lysate for 40 min at 23,710 RCF, 4°C.
33.Mix 5 μl of lysate with 20 μl of 4× protein loading buffer and 55 μl low-salt wash 1 to prepare an electrophoresis sample for the supernatant.
34.Decant the supernatant evenly into two to four new 50-ml Falcon tubes.
35.Add 10 ml of HisPur Ni-NTA resin, equilibrated with lysis buffer, evenly to each tube.
36.Allow the supernatant and Ni resin to gently shake (∼5-10 RPM) for 1 hr at 4°C. Gentle inversion is also acceptable at this point.
37.Spin down the Falcon tubes for 10 min at 3082 RCF, 4°C.
38.Decant the supernatant and label the Ni flowthrough.
39.Prepare an electrophoresis sample for the Ni flowthrough by mixing 5 μl lysate with 20 μl of 4× protein loading buffer and 55 μl low-salt wash 1.
40.Add 40 ml lysis buffer to consolidate the resin into one tube.
41.Spin down the sample in Falcon tubes for 10 min at 3082 RCF, 4°C.
42.Decant the supernatant and label it “wash 1.”
43.Prepare an electrophoresis sample for the wash 1 by mixing 5 μl of wash 1 with 20 μl of 4× protein loading buffer and 55 μl low-salt wash 1.
44.Add 40 ml lysis buffer to transfer the resin to an unpacked plastic column.
45.Wash the resin with 150 ml lysis buffer and label the supernatant “wash 2.”
46.Wash the resin with 50 ml low-salt wash 1 and also collect this supernatant in the “wash 2” container.
47.Prepare an electrophoresis sample for wash 2 by mixing 5 μl of wash 2 with 20 μl of 4× protein loading buffer and 55 μl low-salt wash 1.
48.Elute the sample with 15 ml of the elution buffer 1 three times and label the eluate “elution 1,” “elution 2,” and “elution 3.” Make an electrophoresis sample for the eluted fraction by mixing 20 μl of the eluted sample with 20 μl of 4× protein loading buffer and 40 μl low-salt wash 1.
49.Dilute Ni elution 1 and Ni elution 2 each twofold (1:2) with SPA 1 buffer.
50.Load the diluted Ni elution onto an SP column equilibrated into SPA buffer, using a syringe drive at 2.5 ml/min.
51.Run the SP column at 4°C using the ÄKTA system (flow rate 3 ml/min, 5 ml column) using the following gradients: Wash out unbound protein with 0% SPB 1 buffer (versus SPA 1 buffer) for 5 column volumes; run a linear gradient from 0% SPB 1 to 18% SPB 1 in 5 column volumes; run a linear gradient from 18% SPB 1 to 30% SPB 1 in 20 column volumes; and follow this with a step gradient of 100% SPB 1 for 4 column volumes.
52.Prepare electrophoresis samples for all peaks and run on an SDS-PAGE gel (Fig. 1).
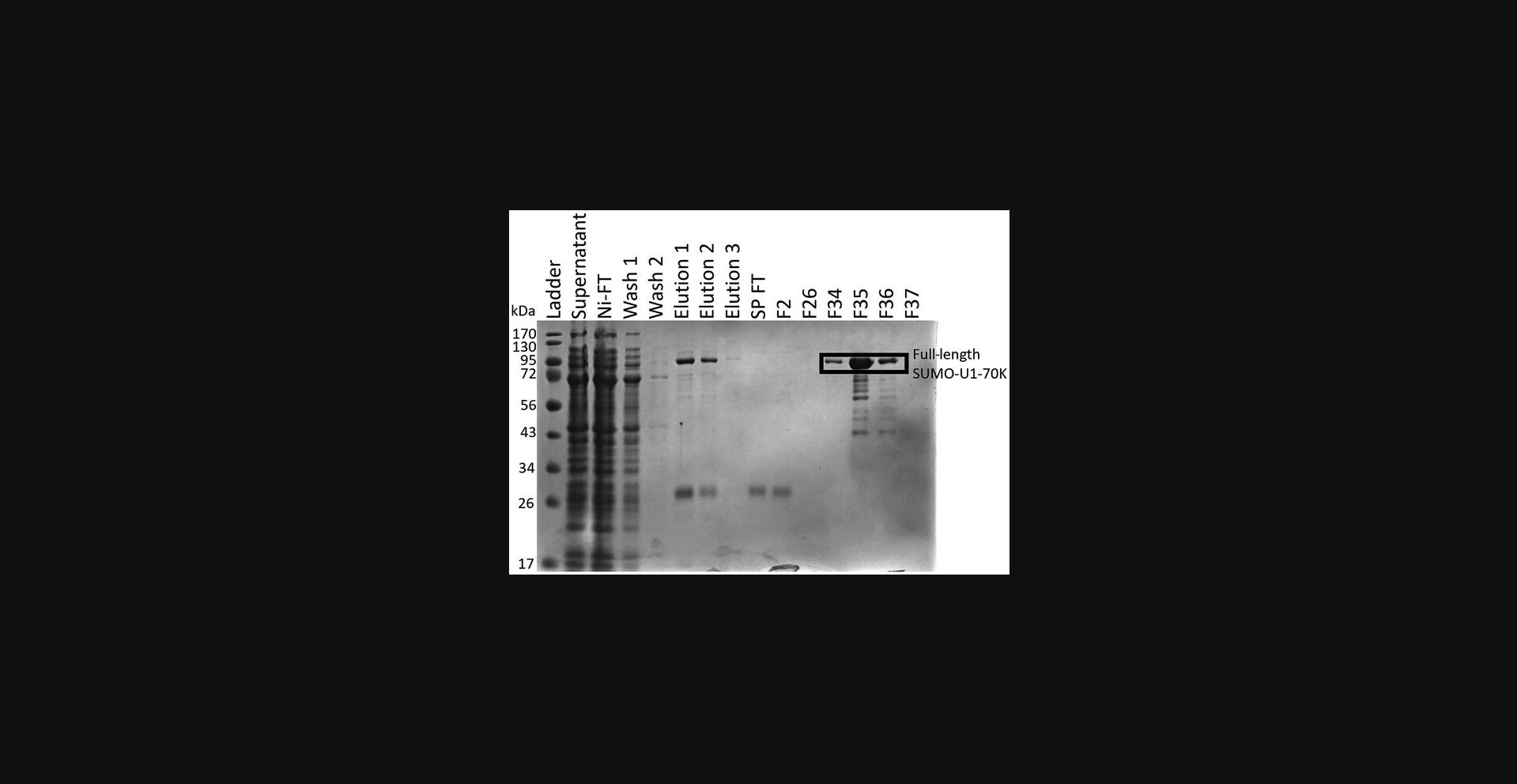
53.Combine fractions containing protein and concentrate using 10-kDa concentrators (Centrifugal Filter Units).
54.Buffer-exchange the protein into storage buffer to remove any urea: Transfer protein to a 10-kDa concentrator and spin the 10-kDa concentrator at 3082 RCF, 4°C, until the volume is ∼1 ml. Then add 1 ml of storage buffer and mix and again spin the 10-kDa concentrator at 3082 RCF, 4°C until the volume is ∼1 ml again. Repeat for a total of 10 times.
55.Store the protein in 1.5-ml low-protein-binding microcentrifuge tubes at −80°C until it is time to use it.
Support Protocol 1: MAKING CHEMICALLY COMPETENT BL21 STAR pRARE/pBB535 CELLS
The BL21 Star cells we used for expression of U1-70K host the pBB535 plasmid, for expression of DnaK and DnaJ, and the pRARE plasmid in commercial Rosetta cells that encodes rare codons. As this cell strain is not commercially available, this protocol describes its preparation.
Materials
-
Rosetta(DE3) Competent Cells (Novagen cat. no. 70953)
-
QIAprep Spin Miniprep Kit (Qiagen, cat. no. 27104)
-
One Shot BL21 Star(DE3) Chemically Competent E. coli (Thermo Fisher Scientific, cat. no. C601003)
-
pBB535 plasmid (Addgene plasmid no. 27392)
-
LB medium (see recipe)
-
LB agar, Miller (Fisher cat. no. BP1425-2)
-
Chloramphenicol (ACROS cat. no. AC227920250) or chloramphenicol stock (see recipe)
-
Spectinomycin (Fisher cat. no. BP29571) or spectinomycin stock (see recipe)
-
Deionized water or Milli-Q water
-
100 mM magnesium chloride (MgCl2; see recipe)
-
100 mm calcium chloride (CaCl; see recipe)
-
85 mm calcium chloride/15% glycerol (see recipe)
-
Ampicillin
-
2-, 10-, 20-, 200-, and 1000-μl pipets
-
10-, 200-, and 1000-µl pipet tips
-
2-, 10-, and 25-ml serological pipets
-
Petri dish (Fisher cat. no. FB087579B)
-
Plate spreader (Fisher cat. no. 14-665-230)
-
Water bath (Fisher CPD02 or equivalent)
-
Incubator shakers for large (New Brunswick Excella E25 or equivalent) and smaller volumes (New Brunswick I 24 or equivalent)
-
Inoculating loops or needles (Fisher cat. no. 22-363-595)
-
Incubator (VWR)
-
NanoDrop (One C) or equivalent spectrophotometer
-
Centrifuges for large (Avanti JXN-26 or equivalent) and small volumes (Allegra X-30R or equivalent)
-
Beckman JA-8000 rotor
-
Centrifuge bottles
-
50-ml conical centrifugal tube (Falcon cat. no. 14-959-49A)
-
1.5-ml low-protein-binding microcentrifuge tubes (ThermoFisher Scientific cat. no. 90410)
-
Liquid nitrogen source
-
−80°C freezer
Day 0: Obtain the pRARE plasmid from Rosetta cells
1.Extract pRARE plasmid from Rosetta cells using QIAprep Spin Miniprep Kit.
Day 1: Transform cells and prepare medium
2.Thaw chemically competent BL21 Star cells on ice.
3.Add 1 μl of pRARE plasmid (from step 1) and 1 μl of pBB535 plasmid to the cells and allow to incubate on ice for 15 min.
4.Heat shock the cells for 60 s at 42°C.
5.Add 150 μl LB to the cells and shake at 37°C for 1 hr.
6.Transfer cells to an agar plate with 50 μg/ml spectinomycin and 50 μg/ml chloramphenicol and spread until dry.
7.Place agar plate at 37°C overnight.
8.Prepare 1.05 L LB medium and put 1 L in a 2.8-L shaker flask and 50 ml in a 250-ml shaker flask.
9.Autoclave LB. Autoclave two 1-L centrifuge bottles and 200 1.5-ml microcentrifuge tubes.
Day 2: Grow cells and induce chemical competence
10.Take agar plate out of incubator (colonies should be visible).
11.Add 50 μg/ml spectinomycin and 50 μg/ml chloramphenicol to the 50 ml of LB medium (starter culture).
12.Pick colonies, use to inoculate the 50-ml LB starter culture, and incubate at 37°C at 220 RPM for 2 hr.
13.After 2 hr, use the starter culture to inoculate the 1 L LB.
14.When the OD600 reaches 0.35-0.4, immediately put the cells on ice. Chill the culture for 20-30 min, swirling occasionally to ensure even cooling. Place centrifuge bottles on ice at this time.
15.Spin 1: Transfer the cultures into ice-cold autoclaved centrifuge bottles. Harvest the cells by centrifugation for 15 min at 3000 × g (∼3470 rpm in Beckman JA-8000 rotor), 4°C.
16.Decant the supernatant and gently resuspend each pellet in ∼200 ml of ice-cold 100 mM MgCl2.
17.Spin 2: Harvest the cells by centrifugation for 15 min at 2000 × g (∼2830 rpm in Beckman JA-8000 rotor), 4°C.
18.Decant the supernatant and resuspend the pellet in ∼200 ml of ice-cold CaCl2. Keep this suspension on ice for at least 20 min. Start placing 1.5-ml microcentrifuge tubes into −80°C freezer if not already chilled.
19.Spin 3: Harvest the cells by centrifugation for 15 min at 2000 × g (∼2830 rpm in the Beckman JA-8000 rotor), 4°C. At this step, put a 50-ml sterile conical tube on ice.
20.Decant the supernatant and resuspend the pellet in ∼50 ml of ice-cold 85 mM CaCl2/15% glycerol. Transfer the suspension to the 50-ml conical tube.
21.Spin 4: Harvest the cells by centrifugation for 15 min at 1000 × g (∼2278 rpm in the Beckman Allegra X30R), 4°C.
22.Decant the supernatant and resuspend the pellet in 4 ml ice-cold 85 mM CaCl2/15% glycerol. The final OD600 of the suspended cells should be ∼200-250.
23.Aliquot 40 μl into sterile 1.5-ml low-protein-binding microcentrifuge tubes and snap freeze with liquid nitrogen. Store frozen cells in the −80°C freezer.
24.Prepare positive and negative controls:
-
Positive control: Confirm the cells are competent and can be transformed with a known plasmid. Transform the cell with a known plasmid and grow them on an agar plate with corresponding antibiotics.
-
Negative control: Confirm the cells are free of contamination by other cell strains, molds, or plasmids. Prepare individual agar plates with 100 μg/ml ampicillin. Spread the cells on these plates. In principle, no colonies should be found. Note that even if there is no plasmid added, it is still necessary to follow the transformation steps.
Basic Protocol 2: PHOSPHORYLATION OF FULL-LENGTH U1-70K USING SRPK1
This protocol describes the phosphorylation of full-length U1-70K with an N-terminal SUMO tag using the SUMO-tagged SRPK1 and its subsequent purification using desalting and SP columns. Typically, the yield of phosphorylated protein is 30%-50% of the input U1-70K.
Materials
-
SUMO-SRPK1 (see Support Protocol 2)
-
SUMO-U1-70K (see Basic Protocol 1)
-
1 M magnesium chloride (MgCl2; see recipe)
-
100 mM ATP (see recipe)
-
Storage buffer (see recipe)
-
Phosphorylation buffer (see recipe)
-
0.5 M EDTA (see recipe)
-
8 M urea (see recipe)
-
Deionized water or Milli-Q water
-
SPA 1 buffer (see recipe)
-
SPB 1 buffer (see recipe)
-
1.5-ml low-protein-binding microcentrifuge tubes (ThermoFisher Scientific cat. no. 90410)
-
2-, 10-, 20-, 200-, and 1000-μl pipets
-
10-, 200-, and 1000-µl pipet tips
-
50-ml conical centrifuge tube (Falcon cat. no. 14-959-49A)
-
15-ml conical centrifuge tube (Thermo Scientific cat. no. 339650)
-
Dialysis tubing (ThermoFisher Scientific cat. no. 68100)
-
2-, 10-, and 25-ml serological pipets
-
Magnetic stir plate and stir bar
-
Incubator: Thermo Scientific Heratherm IMC 18 (VWR) or equivalent
-
HiTrap desalting column (Cytiva cat. no. 17140801)
-
HiTrap SP HP column (Cytiva cat. no. 17115101)
-
ÄKTA Start protein purification system (Cytiva) or equivalent
-
10-kDa spin concentrators (Millipore Sigma UFC901096)
-
Centrifuge (Allegra X-30R or equivalent)
-
NanoDrop (One C) or equivalent spectrophotometer
-
Sonicator (QSONICA Q500)
-
Additional reagents and equipment for SDS-PAGE (Gallagher, 2007)
Day 1
1.Thaw full-length SUMO-SRPK1.
2.Thaw full-length SUMO-U1-70K.
3.Prepare 100 mM ATP and 1 M MgCl2 stock solutions by dissolving solid in deionized water.
4.Determine the volume of U1-70K needed (U).
Example : Stock SUMO-U1-70K = 75 μm, we have 500 l of the stock SUMO-U1-70K, and we plan to phosphorylate all of it. In this example, (U) = 500 μl because we will be phosphorylating all of our sample.
5.Calculate the volume necessary to dilute the U1-70K to 25 μm (FV).
Example :
6.Calculate the volume of ATP needed to make the final concentration 2 mM (A).
Example :
7.Calculate the volume of MgCl2 needed to make the final concentration 10 mM (M).
Example :
8.Calculate the volume of SRPK1 needed to make the final concentration 2.5 μm (SR).
Example : We have 50 μm SUMO-SRPK1 stock:
9.Calculate the volume of storage buffer needed to dilute the stock U1-70K to the final volume (SB).
Example:
10.Mix storage buffer (amount SB), ATP (A), and MgCl2 (M).
Example : Add 880 μl storage buffer, 30 μl of 100 mM ATP, and 15 μl of 1 M MgCl2 to a microcentrifuge tube and mix by pipetting up and down.
11.Add SUMO-SPRK1 (SR) and mix.
Example : Add 75 μl stock SUMO-SRPK1 to the solution from step 10 and mix by pipetting up and down.
12.Add U1-70K (U) and mix, then transfer to 10-kDa dialysis tubing.
Example : Add 500 μl of stock SUMO-U1-70K and mix by pipetting up and down.
13.Multiply FV (Basic Protocol 2, step 5) by 1000 to get the volume of solution that you should use to dialyze the sample (DV).
Example :
14.Mix ATP, MgCl2, storage buffer, and phosphorylation buffer to obtain a buffer with the following composition and volume DV :
- 50 mM Tris·Cl, pH 7.5
- 100 mM Arg/Glu
- 150 mM NaCl
- 0.2 mM TCEP
- 2 mM ATP
- 10 mM MgCl2
Example:
Mix 187.5 ml storage buffer, 1312.5 ml phosphorylation buffer, 30 ml of 100 mM ATP, and 15 ml of 1 M MgCl2 in a beaker.
15.Place a magnetic stir bar in the beaker.
16.Put the dialysis tubing containing the protein from step 12 into the beaker.
17.Incubate at 30°C on a stir plate overnight with slow stirring (at 5-10 rpm).
Day 2
18.Prepare 0.5 M EDTA stock.
19.Calculate the volume of EDTA stock needed:
20.Calculate the volume of 8 M urea stock needed:
21.Mix together EDTA and urea and then add to the sample.
22.Run the sample on a desalting column.
23.Collect fractions containing the protein.
24.Run the 5-ml SP column at 4°C on an ÄKTA protein purification system at a flow rate of 3 ml/min. The protocol should include a wash-out-unbound step with 0% buffer SPB 1 (versus SPA 1) for 5 column volumes (CV), followed by a gradient from 0% buffer SPB 1 to 18% buffer SPB 1 in 5 CV, and a shallow gradient from 18% buffer SPB 1 to 30% buffer SPB c1 in 20 CV. Finally, clean the column using 4 CV of 100% SPB 1.Collect 5-ml fractions during the gradient elution steps.
25.Prepare electrophoresis samples for all peaks and run on an SDS-PAGE (Fig. 3). Phosphorylated protein will come off the column at a lower percentage of buffer SPB 1.
26.Buffer exchange the protein into storage buffer to remove any urea: Transfer protein to a 10-kDa spin concentrator and spin at 3082 RCF, 4°C, until the volume is ∼1 ml. Add 1 ml storage buffer, mix, and spin the 10-kDa concentrator at 3082 RCF, 4°C, until the volume is ∼1 ml again. Repeat process for a total of 10 spins.
Support Protocol 2: PURIFICATION OF SRPK1
This protocol describes the overexpression and purification of active SUMO-tagged SRPK1 for phosphorylation of U1-70K. The typical yield of U1-70K is ∼1 mg per liter of culture.
Materials
-
BL21-CodonPlus(DE3) competent cells (Agilent cat. no. 230245)
-
pSMT3 plasmid encoding human SRPK1 (available from the authors upon request)
-
Luria broth (LB) medium (see recipe)
-
LB agar, Miller (Fisher cat. no. BP1425-2)
-
Kanamycin (Fisher cat. no. 50-213-386) or kanamycin stock (see recipe)
-
Chloramphenicol (ACROS cat. no. AC227920250) or chloramphenicol stock (see recipe)
-
Deionized water or Milli-Q water
-
Loading buffer (see recipe)
-
Elution buffer 4 (see recipe)
-
Buffer QA (see recipe)
-
Buffer QB (see recipe)
-
IPTG (Fisher cat. no. BP1755-100)
-
4× protein loading buffer (Gallagher, 2007)
-
HisPur Ni-NTA resin (Thermo Scientific cat. no. 88222)
-
Water bath (Fisher CPD02 or equivalent)
-
2-, 10-, 20-, 200-, and 1000-μl pipets
-
10-, 200-, and 1000-µl pipet tips
-
2-, 10-, and 25-ml serological pipets
-
Petri dish (Fisher cat. no. FB087579B)
-
Plate spreader (Fisher cat. no. 14-665-230)
-
2.8-L and 250-ml culture flasks
-
Inoculating loops or needles (Fisher cat. no. 22-363-595)
-
Incubator (VWR)
-
Incubator shaker (New Brunswick Excella E25 or equivalent)
-
Incubator shaker (New Brunswick I 24 or equivalent)
-
NanoDrop (One C) or equivalent spectrophotometer
-
Centrifuge (Avanti JXN-26 or equivalent)
-
Centrifuge (Allegra X-30R or equivalent)
-
Centrifuge (Sorvall Legend X1R or equivalent)
-
Beckman JA-8000 rotor
-
Centrifuge bottles
-
−80°C freezer
-
Sonicator (QSONICA Q500)
-
5-ml HiTrap Q HP column (Cytiva)
-
ÄKTA Start protein purification system (Cytiva) or equivalent
-
10-kDa spin concentrators (Millipore Sigma UFC901096)
-
1.5-ml low-protein-binding microcentrifuge tubes (ThermoFisher Scientific cat. no. 90410)
-
50-ml conical centrifugal tube (Falcon cat. no. 14-959-49A)
-
15-ml conical centrifugal tube (Thermo Scientifics cat. no. 339650)
-
Additional reagents and equipment for SDS-PAGE (Gallagher, 2007)
Day 1: Transform and Culture Cells
Transform competent cells and spread on plate
1.Thaw chemically competent BL21-CodonPlus cells on ice.
2.Add 1 μl of pSMT3-SRPK1 plasmid to the cells and incubate cells on ice for 15 min.
3.Heat-shock the cells for 60 s at 42°C using a water bath.
4.Add 150 μl LB to the cells and allow to shake for 1 hr at 37°C.
5.Transfer cells to an agar plate with 50 μg/ml kanamycin and 50 μg/ml chloramphenicol. Spread until dry.
6.Incubate plate overnight at 37°C.
Prepare medium and buffers
7.Prepare 6.05 L of LB.
8.Add 1 L LB to each of six clean 2.8-L culture flasks and 50 ml LB to one 250-ml culture flask.
9.Autoclave LB.
10.Add antibiotics (50 μg/ml kanamycin and 50 μg/ml chloramphenicol) to the 50 ml LB (starter culture) after it has cooled.
11.Make buffers: loading buffer, elution buffer 4, buffer QA, and buffer QB.
Day 2: Prepare Starter Culture
12.Pick colonies from the agar plate with inoculating loops and use to inoculate the 50-ml starter culture.
13.Incubate starter culture with shaking at 37°C overnight. Below step 7 (Support Protocol 2), please add the following as a separate italic “annotation” paragraph:To avoid contamination, do not prepare cell culture media more than 1-2 days before they are needed.
Day 3: Grow and Induce Cells
14.Check the OD600 of the starter culture.
15.Calculate how much starter culture is needed to add to 1 L of LB to obtain a starting OD600 of 0.02.
16.Add antibiotics (50 μg/ml kanamycin and 50 μg/ml chloramphenicol) to each of the culture flasks containing 1 L of LB from step 8.
17.Inoculate the LB with the volume of starter calculated in step 15.
18.Grow at 37°C until culture reaches an OD600 of 0.6-0.8 (∼8 hr).
19.Add 500 μl of 1 M IPTG to each flask.
20.Reduce the temperature to 22°C and allow the cells to shake overnight (16 hr).
Day 4: Protein Harvest and Purification
Harvest protein
21.Collect cells using Beckman JA-8000 rotor at 3000 RCF for 15 min at 4°C.
22.Check the mass of cells collected.
23.Freeze the cell pellet.
Purify protein
24.Thaw cells.
25.Resuspend cells with 4 ml loading buffer per gram of cells.
26.Freeze/thaw cells twice: Incubate the sample in a −80°C freezer for 20 min to freeze, and then incubate the sample in a room-temperature water bath until cells have thawed (∼5 min). Repeat the procedure.
27.Thaw the cells.
28.Sonicate cells (in a metal or plastic beaker) using an ice-water bath to keep them cool: Set sonicator to 70% power, pulsing for 3 s on and 5 s off, and sonicate for three cycles each comprising a total of 1 min of sonication, allowing the sample to cool for 5 min between rounds.
29.Spin down cell lysate for 40 min at 23,710 RCF, 4°C.
30.Prepare an electrophoresis sample for the supernatant by mixing 5 μl lysate with 20 μl of 4× protein loading buffer and 55 μl low-salt wash.
31.Load supernatant onto 5 ml of HisPur Ni-NTA resin equilibrated with loading buffer and label flowthrough “Ni flowthrough.”
32.Prepare an electrophoresis sample for the Ni flowthrough by mixing 5 μl Ni flowthrough with 20 μl of 4× protein loading buffer and 55 μl low-salt wash.
33.Wash resin with 200 ml loading buffer and label the flowthrough “high-salt wash.”
34.Prepare an electrophoresis sample for the high-salt wash by mixing 5 μl high-salt wash with 20 μl of 4× protein loading buffer and 55 μl low-salt wash.
35.Wash resin with 50 ml buffer QA and label the flowthrough “low-salt wash.”
36.Prepare an electrophoresis sample for the low-salt wash by mixing 5 μl lysate with 20 μl of 4× protein loading buffer and 55 μl low-salt wash.
37.Elute the sample with 15 ml elution buffer 4 twice, and label the eluates “elution 1” and “elution 2.” Prepare an electrophoresis sample for the eluted fraction by mixing 20 μl of the eluted sample with 20 μl of 4× protein loading buffer and 40 μl low-salt wash.
38.Dilute the Ni elution twofold (1:2) with buffer QA.
39.Load the diluted Ni elution onto an equilibrated Q column using a syringe drive at 2.5 ml/min.
40.Run the Q column on an ÄKTA system at room temperature, with a CV of 5 ml and a flow rate of 3 ml/min, and with the following gradient: Wash out unbound protein with 5 CV of 0% buffer QB, and then run a linear gradient from 0% buffer QB to 100% buffer QB in 25 column volumes. Set the fraction volume to 5 ml.
41.Prepare electrophoresis samples for all peaks and run on an SDS-PAGE gel (Fig. 4). Phosphorylated protein will come off the column at a lower percentage of buffer QB.
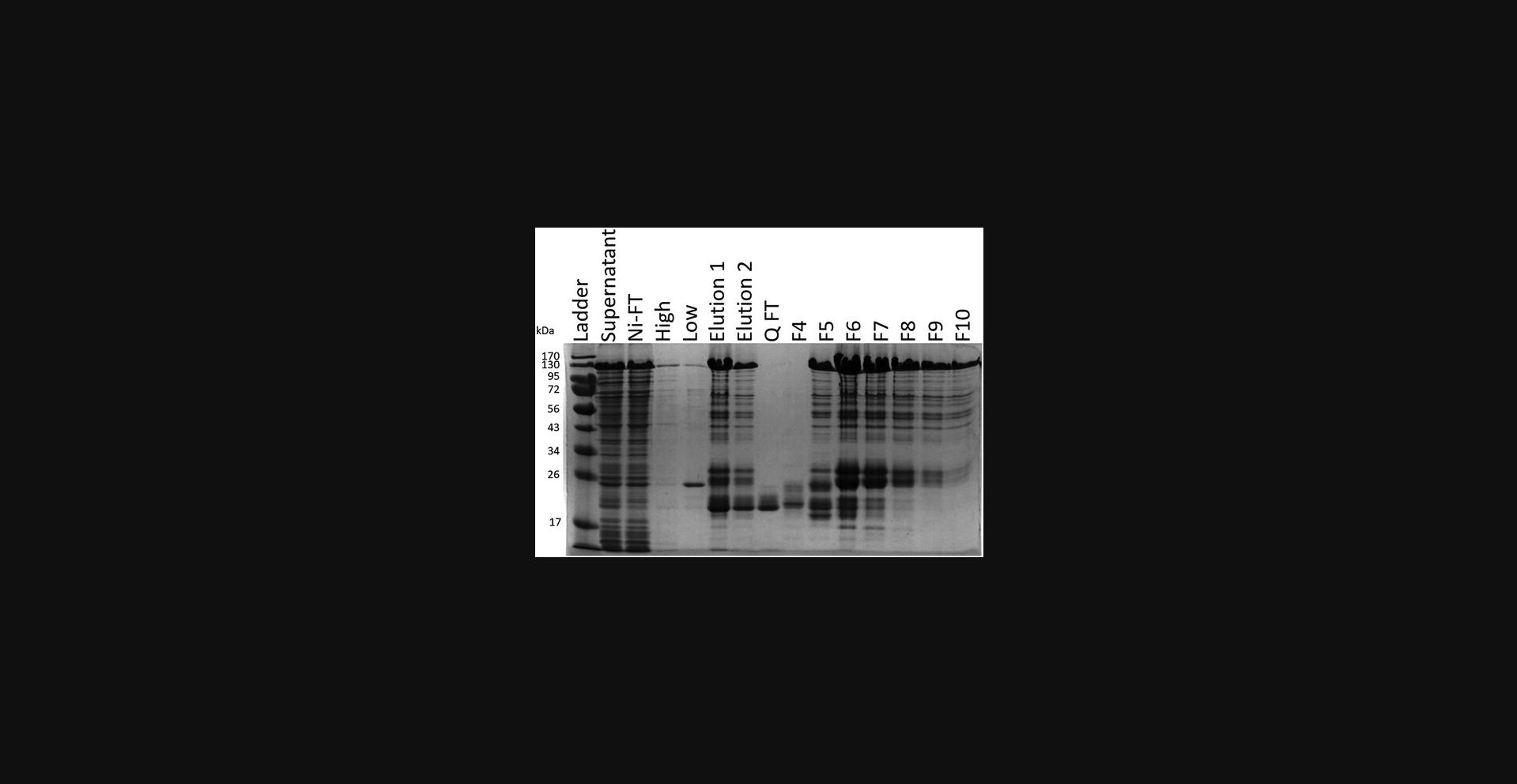
42.Combine and concentrate fractions containing protein using a 10-kDa spin concentrator. Store frozen at −80°C in 1.5-ml low-protein-binding microcentrifuge tubes.
Basic Protocol 3: EXPRESSION AND PURIFICATION OF U1-70K BAD1 FROM E. coli
This protocol describes the overexpression and purification of U1-70K's BAD1 domain using BL21-CodonPlus(DE3) cells with pBB535.
Materials
-
BL21-CodonPlus(DE3) competent cells (Agilent cat. no. 230245)
-
pSMT3 plasmid encoding U1-70K 229-306+W (codon optimized; available from the authors upon request)
-
pBB535 plasmid (Addgene plasmid no. 27392)
-
Deionized water or Milli-Q water
-
Luria broth (LB) medium (see recipe)
-
LB agar, Miller (Fisher cat. no. BP1425-2)
-
Kanamycin (Fisher cat. no. 50-213-386)
-
Chloramphenicol (ACROS cat. no. AC227920250) or chloramphenicol stock (see recipe)
-
Spectinomycin (Fisher cat. no. BP29571) or spectinomycin stock (see recipe)
-
Terrific broth (TB) medium (see recipe)
-
Lysis buffer 1 (see recipe)
-
Low-salt wash 2 (see recipe)
-
Elution buffer 2 (see recipe)
-
SPA 2 (see recipe)
-
SPB 2 (see recipe)
-
IPTG (Fisher cat. no. BP1755-100)
-
4× protein loading buffer (Gallagher, 2007)
-
HisPur Ni-NTA resin (Thermo Scientific cat. no. 88222)
-
Upl1 (SUMO protease; Thermo Fisher Scientific cat. no. 12588018)
-
1 M Tris·Cl, pH 8.0 (see recipe)
-
Water bath (Fisher cat. no. CPD02 or equivalent)
-
2.8-L and 250-ml culture dishes
-
Petri dish (Fisher cat. no. FB087579B)
-
Inoculating loops or needles (Fisher cat. no. 22-363-595)
-
Plate spreader (Fisher cat. no. 14-665-230)
-
2-, 10-, 20-, 200-, and 1000-μl pipets
-
10-, 200-, and 1000-µl pipet tips
-
2-, 10-, and 25-ml serological pipets
-
Incubator shaker (New Brunswick Excella E25 or equivalent)
-
Incubator shaker (New Brunswick I 24 or equivalent)
-
Incubator (VWR)
-
NanoDrop (One C) or equivalent spectrophotometer
-
Centrifuges: Avanti JXN-26, Allegra X-30R, and Sorvall Legend X1R, or equivalent
-
Beckman JA-8000 rotor
-
Centrifuge bottles
-
50-ml conical centrifugal tube (Falcon cat. no. 14-959-49A)
-
15-ml conical centrifugal tube (Thermo Scientific cat. no. 339650)
-
−80°C freezer
-
Sonicator (QSONICA Q500)
-
5-ml HiTrap SP HP column (Cytiva cat. no. 17115101)
-
ÄKTA Start protein purification system (Cytiva) or equivalent
-
3-kDa spin concentrators (Millipore Sigma cat. no. UFC900396)
-
1.5-ml low-protein-binding microcentrifuge tubes (ThermoFisher Scientific cat. no. 90410)
-
Additional reagents and equipment for SDS-PAGE (Gallagher, 2007)
Day 1
Transform the competent cells and spread on the plate
1.Thaw chemically competent BL21-CodonPlus(DE3) cells on ice.
2.Add 1 μl of pSMT3-U1-70K BAD1 plasmid and 1 μl of pBB535 plasmid to the cells, and incubate on ice for 15 min.
3.Heat shock the cells for 60 s at 42°C.
4.Add 150 μl of LB to the cells and shake for 1 hr at 200 RPM, 37°C.
5.Transfer cells to an agar plate with 50 μg/ml kanamycin, 50 μg/ml chloramphenicol, and 50 μg/ml spectinomycin. Spread until dry.
Prepare medium
6.Prepare 6.05 L of TB.
7.Add 1 L TB to each of six clean 2.8-L culture flasks and 50 ml TB to one 250-ml culture flask.
8.Autoclave TB.
Day 2
Prepare starter culture
9.Add 50 μg/ml kanamycin, 50 μg/ml chloramphenicol, and 50 μg/ml spectinomycin to the 50 ml TB (starter culture).
10.Scrape colonies off the plate and use these to inoculate the 50-ml starter culture.
11.Incubate the starter culture overnight at 37°C with shaking at 220 RPM.
Prepare buffers
12.Prepare and filter all buffers (lysis buffer, low-salt wash 2, elution buffer 2, SPA 2, and SPB 2).
Day 3: Grow and induce the cells
13.Check the OD600 of the starter culture.
14.Calculate how much starter culture needs to be added to 1 L of TB to make the starting OD600 0.02.
15.Add 50 μg/ml kanamycin, 50 μg/ml chloramphenicol, and 50 μg/ml spectinomycin to each of the flasks containing 1 L of TB.
16.Inoculate the 1 L TB with the volume of starter culture calculated in step 14.
17.Incubate at 37°C until the culture reaches an OD600 of 0.8-1.0.
18.Add 500 μl of 1 M IPTG to each flask.
19.Reduce the temperature to 25°C, and incubate the cells overnight with shaking at 220 RPM.
Day 4: Harvest cells
20.Collect cells by centrifugation for 15 min using a Beckman JA-8000 rotor at 3000 RCF, 4°C.
21.Check mass of the cells collected.
22.Freeze cell pellet.
Day 5: Purify the protein
23.Thaw cells.
24.Resuspend cells with 1.5 ml of lysis buffer per gram of cells.
25.Freeze-thaw cells twice.
26.Sonicate cells using an ice-water bath to keep cool: Set the sonicator to 70% power, pulsing for 3 s on and 5 off, and sonicate for three to six cycles of 1 min each, allowing the sample to cool for 5 min between rounds.
27.Spin down the cell lysate for 40 min at 23,710 RCF, 4°C.
28.Mix 5 μl of the supernatant with 20 μl of 4× protein loading buffer and 55 μl low-salt wash to prepare an electrophoresis sample for the supernatant.
29.Place 5 ml of Ni resin (unpacked) in an empty 5-ml column and equilibrate with lysis buffer. Apply the supernatant to the column.
30.Mix 5 μl of the flowthrough with 20 μl of 4× protein loading buffer and 55 μl low-salt wash to prepare an electrophoresis sample for the Ni flowthrough.
31.Wash the Ni resin with 200 ml lysis buffer.
32.Mix 5 μl of the flowthrough from this step with 20 μl of 4× protein loading buffer and 55 μl low-salt wash to prepare an electrophoresis sample for the high-salt wash.
33.Wash the resin with 50 ml low-salt wash.
34.Elute the sample with 15 ml of elution buffer 4 times and label “elution 1,” “elution 2,” “elution 3,” and “elution 4.” Make an electrophoresis sample for the eluted fractions by mixing 20 μl of the eluted sample with 20 μl of 4× protein loading buffer and 40 μl low-salt wash.
35.Dilute Ni elutions 1-3 each twofold (1:2) with SPA buffer.
36.Load the diluted Ni elution onto an equilibrated SP column using a syringe drive at 2.5 ml/min.
37.Run the 5-ml SP column on an ÄKTA system at room temperature. Start with a gradient from 0% to 40% buffer SPB 2 in 6 CV, followed by a gradient from 40% to 60% buffer SPB 2 in 19 CV. Finally, run 100% buffer SPB 2 through the column for 5 CV. Collect 5-ml fractions throughout the protocol.
38.Run samples of the fractions on an SDS-PAGE gel to confirm which fractions contain the protein.
39.Add 200 μl of 1 M Tris·Cl, pH 8.0, to each 5-ml fraction containing pure BAD1 to adjust the pH to ∼7.0.
40.Combine fractions and add 200 μl of 12.5 μM Ulp1.
41.Incubate samples for 2 hr at 37°C.
42.Dilute sample twofold (1:2) with SPA 2 buffer.
43.Load sample onto an equilibrated SP column using a syringe drive at 2.5 ml/min.
44.Run an SP column on an ÄKTA system at room temperature, with a gradient from 0% to 40% buffer SPB 2 in 6 CV, followed by a gradient from 40% to 60% buffer SPB 2 in 19 CV. Finally, wash with 100% buffer SPB 2 for 5 CV.
45.Prepare electrophoresis samples for all peaks and run on an SDS-PAGE gel (Fig. 5). Fractions containing BAD1 are identified based on size and the percentage of buffer SPB 2 in the fractions.
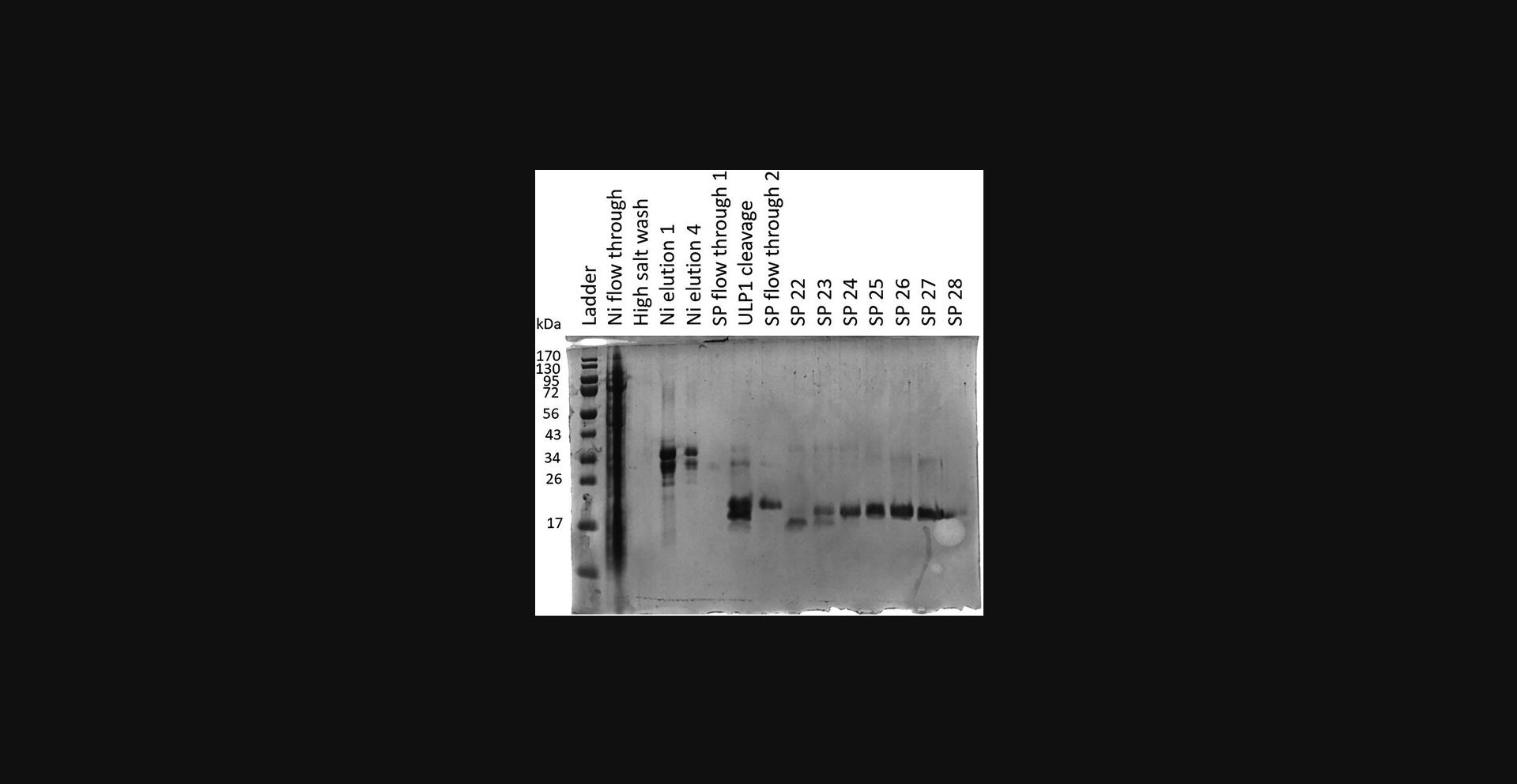
46.Combine fractions containing pure protein, concentrate using a 3-kDa spin concentrators (Centrifugal Filter Units), and buffer exchange into a lower-salt buffer (as convenient; a high-salt buffer is not required). Store frozen at −80°C in 1.5-ml low-protein-binding microcentrifuge tubes.
Basic Protocol 4: PHOSPHORYLATION OF U1-70K BAD1 USING SRPK1
This protocol describes the phosphorylation of U1-70K's BAD1 domain using SUMO-tagged SRPK1 and the subsequent purification using a Mono S column.
Materials
-
SUMO-SRPK1 (see Support Protocol 2)
-
U1-70K BAD1+W (see Basic Protocol 3)
-
Deionized water or Milli-Q water
-
Phosphorylation buffer (see recipe)
-
1 M magnesium chloride (see recipe)
-
100 mM ATP (see recipe)
-
0.5 M EDTA (see recipe)
-
SPA 2 buffer (see recipe)
-
SPB 2 buffer (see recipe)
-
2-, 10-, 20-, 200-, and 1000-μl pipets
-
10-, 200-, and 1000-µl pipet tips
-
2-, 10-, and 25-ml serological pipets
-
50-ml conical centrifugal tube (Falcon cat. no. 14-959-49A)
-
15-ml conical centrifugal tube (Thermo Scientific cat. no. 339650)
-
Centrifuge (Allegra X-30R or equivalent)
-
1.5-ml low-protein-binding microcentrifuge tubes (ThermoFisher Scientific cat. no. 90410)
-
Incubator (VWR)
-
Thermo Scientific Heratherm IMC 18 or equivalent
-
Mono S column (Cytiva)
-
ÄKTA Start protein purification system (Cytiva) or equivalent
-
Centrifugal Filter Units (Amicon® Ultra-15 or 4)
-
NanoDrop (One C) or equivalent spectrophotometer
-
3-kDa spin concentrators (Millipore Sigma UFC900396)
-
Additional reagents and equipment for SDS-PAGE (Gallagher, 2007)
1.Thaw full-length SUMO-SRPK1.
2.Thaw BAD1.
3.Buffer-exchange the protein into phosphorylation buffer: Transfer protein to a 3-kDa concentrator. Spin the 3-kDa concentrator at 3082 RCF, 4°C, until the volume is ∼1 ml or less. Add 1 ml phosphorylation buffer, mix, and spin the 10-kDa concentrator at 3082 RCF, 4°C, until the volume is ∼1 ml again. Repeat for a total of 10 times.
4.Make 100 mM ATP and 1 M MgCl2 stock solutions.
5.Determine the volume of the BAD1 sample to use (= U):
Example : We have 1000 μl of 500 μm BAD1 and we intend to phosphorylate all of it, so in this case U = 1000 μl.
6.Calculate the volume necessary to dilute the BAD1 sample to 100 μm (= FV):
Example :
7.Calculate the volume of ATP needed to make the final concentration 2 mM (= A):
Example :
8.Calculate the volume of MgCl2 needed to make the final concentration 10 mM (= M):
Example :
9.Calculate the volume of SRPK1 needed to make the final concentration 1 μm (= SR):
Example : We have 100 μm SRPK1 stock:
10.Calculate the volume of phosphorylation buffer needed (=
Example :
11.Add phosphorylation buffer (P) to a Falcon tube and mix by inverting.
Example : Add 3800 μl phosphorylation buffer to a 15-ml Falcon tube.
12.Add ATP (A) and MgCl2 (M) to the tube and mix.
Example : Add 100 μl of 100 mM ATP and 50 μl of 1 M MgCl2.
13.Add SRPK1 stock (SR) to the tube and mix by inverting.
Example : Add 50 μl of 100 μm SRPK1 stock.
14.Add BAD1 stock (U) to the tube and mix.
Example : Add 1000 μl of 500 μm BAD1.
15.Incubate 1 hr at 30°C.
16.Add 0.5 M EDTA to the sample to make the final concentration 15 mM EDTA.
17.Incubate the sample 15 min at 25°C with shaking at 10 RPM.
18.Dilute the sample threefold (1:3) with SPA 2 buffer.
19.Load the sample onto a Mono S column.
20.Run the Mono S column at 4°C on the ÄKTA system using a gradient from 0% to 100% SPB 2 (versus SPB A) for 100 column volumes with a fractionation volume of 1.5 ml.
21.Prepare electrophoresis samples for all peaks and run on an SDS-PAGE gel (Fig. 6).
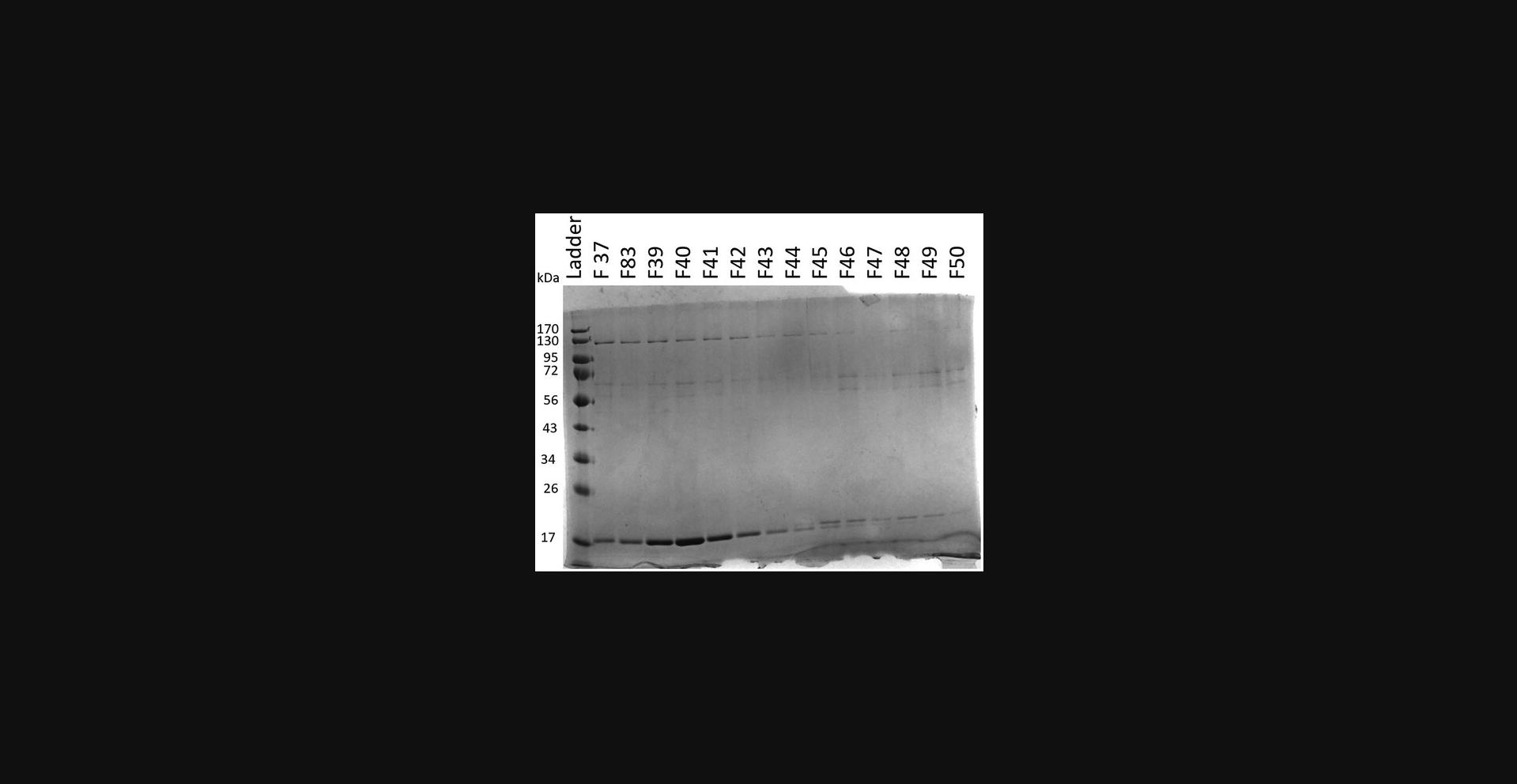
22.Combine fractions containing phosphorylated BAD1 and concentrate using 3-kDa spin concentrator.
23.Store protein at −80°C.
Basic Protocol 5: EXPRESSION AND PURIFICATION OF U1-70K BAD2 FROM E. coli
This protocol describes the overexpression and purification of the U1-70K BAD2 domain using BL21 Star pRARE cells.
Materials
-
BL21 Star pRARE/cells, chemically competent (see Support Protocol 1)
-
pSMT3 plasmid encoding the U1-70K 348-390+W (codon optimized; available from the authors upon request)
-
Lysogeny broth (LB) medium (see recipe)
-
LB agar, Miller (Fisher cat. no. BP1425-2)
-
Kanamycin (Fisher cat. no. 50-213-386) or kanamycin stock (see recipe)
-
Chloramphenicol (ACROS cat. no. AC227920250) or chloramphenicol stock (see recipe)
-
Spectinomycin (Fisher cat. no. BP29571) or spectinomycin stock (see recipe)
-
Terrific broth (TB) medium (see recipe)
-
Pierce protease inhibitor mini tablets (Thermo Scientific cat. no. A32953)
-
Deionized water or Milli-Q water
-
Lysis buffer (see recipe)
-
Low-salt wash 2 (see recipe)
-
Elution buffer 3 (see recipe)
-
SPA 2 (see recipe)
-
SPB 2 (see recipe)
-
IPTG (Fisher cat. no. BP1755-100)
-
4× protein loading buffer (Gallagher, 2007)
-
HisPur Ni-NTA resin (Thermo Scientific cat. no. 88222)
-
Upl1 (SUMO protease; Thermo Fisher Scientific cat. no. 12588018)
-
Small protein ladder (Invitrogen cat. no. LC5925)
-
Tricine (Sigma-Aldrich EC cat. no. 227-193-6)
-
Water bath (Fisher cat. no. CPD02 or equivalent)
-
Petri dish (Fisher cat. no. FB087579B)
-
Plate spreader (Fisher cat. no. 14-665-230)
-
Inoculating loops or needles (Fisher cat. no. 22-363-595)
-
2-, 10-, 20-, 200-, and 1000-μl pipets
-
10-, 200-, and 1000-µl pipet tips
-
2-, 10-, and 25-ml serological pipets
-
2.8-L and 50-ml culture flasks
-
NanoDrop (One C) or equivalent spectrophotometer
-
Incubator shaker (New Brunswick Excella E25 or equivalent)
-
Incubator shaker (New Brunswick I 24 or equivalent)
-
Incubator (VWR)
-
Centrifuge (Avanti JXN-26 or equivalent)
-
Centrifuge (Allegra X-30R or equivalent)
-
Centrifuge (Sorvall Legend X1R or equivalent)
-
Beckman JA-8000 rotor
-
Centrifuge bottles
-
−80°C freezer
-
50-ml conical centrifugal tube (Falcon cat. no. 14-959-49A)
-
15-ml conical centrifugal tube (Thermo Scientifics cat. no. 339650)
-
Sonicator (QSONICA Q500)
-
5-ml HiTrap SP HP column (Cytiva, cat. no. 17115101)
-
ÄKTA Start protein purification system (Cytiva) or equivalent
-
3-kDa spin concentrators (Millipore Sigma cat. no. UFC900396)
-
Additional reagents and equipment for SDS-PAGE (Gallagher, 2007)
Day 1
Transform competent cells and spread on plate
1.Thaw BL21 Star pRARE cells on ice.
2.Add 1 μl pSMT3 U1-70K BAD2 plasmid and incubate on ice for 15 min.
3.Heat shock cells at 42°C for 1 min.
4.Add 150 μl LB medium to the cells and allow them to shake for 1 hr.
5.Transfer cells to an agar plate with 50 μg/ml kanamycin, 50 μg/ml chloramphenicol, and 50 μg/ml spectinomycin antibiotics.
6.Allow the agar plate to incubate at 37°C overnight.
Prepare medium
7.Prepare 6.05 L of TB.
8.Place 1 L TB into each of six 2.8-L culture flasks and 50 ml TB in a 250-ml culture flask.
9.Autoclave medium.
Day 2
10.Add antibiotics (50 μg/ml kanamycin, 50 μg/ml chloramphenicol, and 50 μg/ml spectinomycin) to the 50 ml TB (starter culture).
11.Remove the agar plate from the incubator.
12.Scrape cells from the agar plate and add to the starter culture.
13.Shake the starter culture at 37°C overnight.
14.Prepare buffers: lysis buffer, low-salt wash 2, elution buffer 3, SPA 2, and SPB 2.
Day 3
15.Check the OD600 of the starter culture.
16.Calculate how much starter culture needs to be added to 1 L TB to make the starting OD600 0.02.
17.Add antibiotics (50 μg/ml kanamycin and 50 μg/ml chloramphenicol) to each of the culture flasks containing 1 L TB.
18.Inoculate each flask with the volume of starter calculated in step 15.
19.Grow cultures at 37°C until they reach an OD600 of 0.8-1.0.
20.Add 500 μl of 1 M IPTG to each flask.
21.Reduce the temperature to 25°C and allow the cells to shake overnight at 220 RPM.
Day 4
Harvest cells
22.Collect cells using Beckman JA-8000 rotor for 15 min at 3000 RCF, 4°C.
23.Check the mass of cells collected.
24.Freeze the cell pellet.
Purify the protein
25.Thaw cells.
26.Resuspend cells with 1.5 ml lysis buffer per gram of cells. Freeze/thaw cells twice: Incubate the sample in a −80°C freezer for 20 min to freeze, then incubate the sample in a room-temperature water bath until cells have thawed (∼5 min), and repeat the procedure.
27.Sonicate cells in an ice bath: Set the sonicator to 70% power, pulsing for 3 s on and 5 s off, and sonicate for three cycles of 1 min each, allowing the sample to cool down for 5 min between rounds of sonication.
28.Spin down cell lysate for 40 min at 23,710 RCF, 4°C.
29.Prepare an electrophoresis sample for the supernatant by mixing 5 μl lysate with 20 μl of 4× protein loading buffer and 55 μl low-salt wash.
30.Apply supernatant to 5 ml unpacked Ni resin equilibrated with lysis buffer.
31.Prepare an electrophoresis sample for the Ni flowthrough by mixing 5 μl lysate with 20 μl of 4× protein loading buffer and 55 μl low-salt wash.
32.Wash Ni resin with 200 ml lysis buffer.
33.Prepare an electrophoresis sample for the high-salt wash by mixing 5 μl lysate with 20 μl of 4× protein loading buffer and 55 μl low-salt wash.
34.Wash resin with 50 ml low-salt wash.
35.Prepare an electrophoresis sample for the low-salt wash by mixing 5 μl lysate with 20 μl of 4× protein loading buffer and 55 μl low-salt wash.
36.Elute the sample with 15 ml elution buffer 3 three times and label the eluates elution 1, elution 2, and elution 3.Make an electrophoresis sample for each eluted fraction by mixing 20 μl of the eluted sample with 20 μl of 4× protein loading buffer and 40 μl low-salt wash.
37.Add Ulp1 to the Ni elution and incubate the sample at 37°C for 1 hr.
38.Dilute the sample threefold (1:3) with SPA buffer.
39.Load the diluted Ni elution onto an equilibrated SP column at a flow rate of 3 mL/min using a syringe drive at 2.5 ml/min.
40.Run a 5-ml SP column on an ÄKTA system at room temperature with a gradient from 0% to 100% in 25 CV while collecting 5-ml fractions.
41.Prepare electrophoresis samples for all peaks and run an SDS-PAGE gel (Fig. 7).
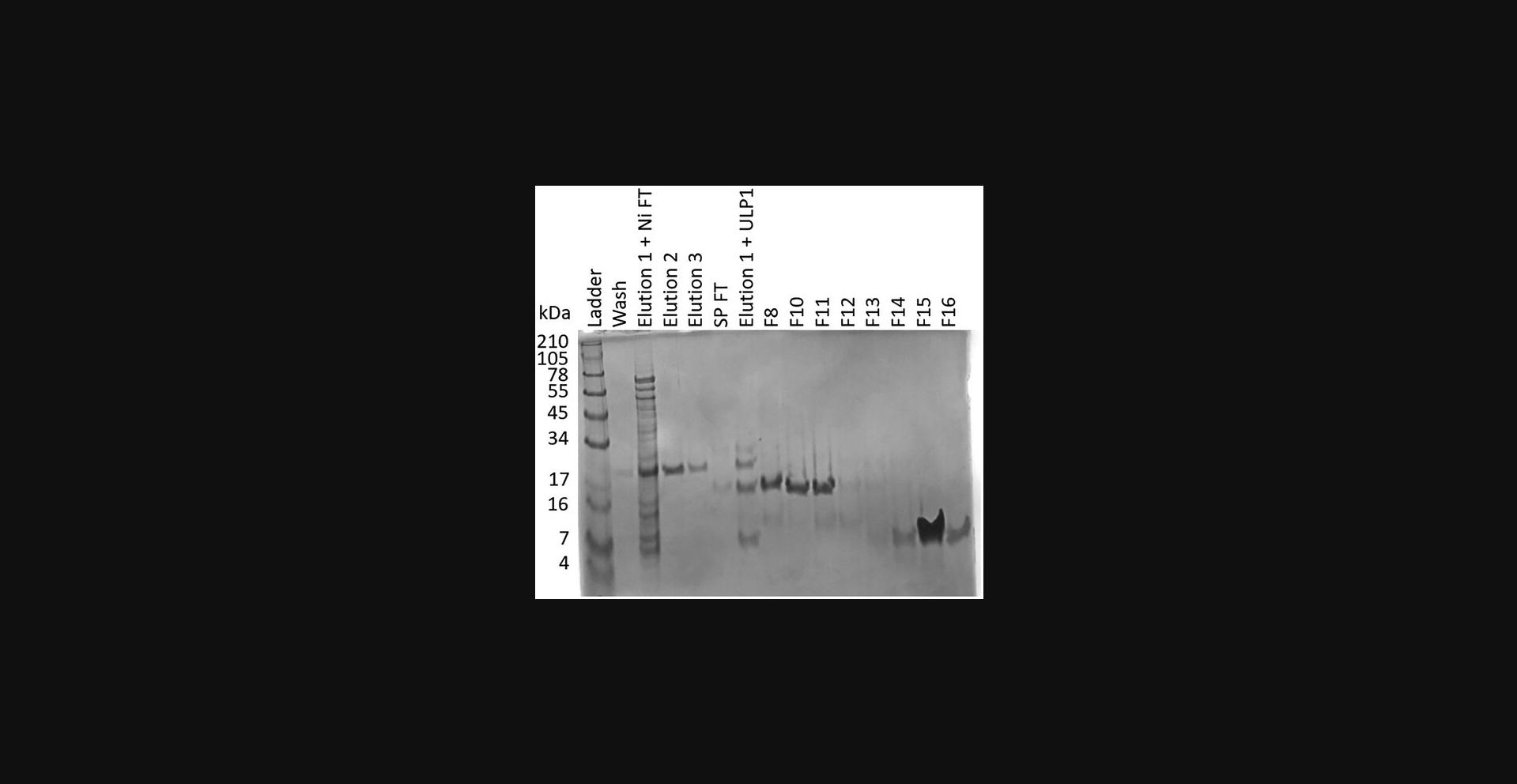
42.Combine fractions containing protein and concentrate using a 3-kDa spin concentrator (Centrifugal Filter Unit).
43.Label and store the protein in 1.5-ml low-protein-binding microcentrifuge tubes at −80°C until ready for use.
REAGENTS AND SOLUTIONS
Arg/Glu, 1 M, with varying pH (1 L)
- 174.2 g arginine (Thermo Scientific cat. no. 104990025)
- Dissolve in 700 ml of distilled, deionized water (ddH2O) Disso or Milli-Q water
- Add glutamate to desired pH (ACROS cat. no. 156210010)
- Add 1 ml of 20% NaN3 (see recipe)
- Bring volume to 1 L with ddH2O or Milli-Q water
- Sterilize by filtration using a 0.22-μm membrane filter (Thermo Scientific cat. no. 50-109-8762)
- Store up to 2 months at room temperature or 4°C
ATP (adenosine 5′-triphosphate disodium salt hydrate), 0.1 M (100 ml)
- 5.51 g ATP (Thermo Scientific cat. no. AAL1452203)
- Dissolve in 100 ml ddH2O or Milli-Q water
- Sterilize by filtration using a 0.22-μm membrane filter (Thermo Scientific cat. no. 50-109-8762)
- Store up to 2 months at −20°C
Buffer QA (1 L)
- 20 ml 1 M HEPES-NaOH, pH 7.5
- 0.2 ml 1 M TCEP (see recipe)
- Fill to final volume with ddH2O or Milli-Q water
- Filter using a 0.22-μm filter membrane.
- Store at 4°C
Final: 20 mM HEPES-NaOH, pH 7.5, 0.2 mM TCEP
Buffer QB (1 L)
- 20 ml 1 M HEPES-NaOH, pH 7.5
- 0.2 ml 1 M TCEP (see recipe)
- 400 ml 5 M NaCl (see recipe)
- Fill to final volume with ddH2O or Milli-Q water
- Filter using a 0.22-μm filter membrane
- Store at 4°C
Final: 20 mM HEPES-NaOH, pH 7.5, 2 M NaCl, 0.2 mM TCEP.
Calcium chloride, 100 mM (1000 ml)
- 11.098 g calcium chloride (Thermo Fisher Scientific cat. no. L13191.0B)
- Dissolve in 1000 ml ddH2O or Milli-Q water
- Sterilize by filtration using a 0.22-μm membrane filter (Thermo Scientific cat. no. 50-109-8762)
- Store up to 12 months at 4°C
Calcium chloride/15% glycerol (1000 ml)
- 9.433 g calcium chloride (Thermo Fisher Scientific cat. no. L13191.0B)
- Dissolve in 850 ml ddH2O or Milli-Q water
- Add 150 ml glycerol (EC No. 200-289-5)
- Sterilize by filtration using a 0.22-μm membrane filter (Thermo Scientific cat. no. 50-109-8762)
- Store up to 12 months at 4°C
Chloramphenicol stock, 50 mg/ml, 1000× (10 ml)
- 0.5 g chloramphenicol (ACROS cat. no. AC227920250)
- Dissolve in 10 ml ethanol
- Sterilize using a syringe filter unit (Millipore cat. no. SLGV033RS)
- Store up to 6 months at 4°C
EDTA, 0.5 M (10 ml)
- Dissolve 1.86 g disodium EDTA (Fisher cat. no. BP120-1) in 8 ml Milli-Q water
- Use 10 M NaOH solution to adjust pH to 8.0
- Add more Milli-Q water to 10 ml final volume
- Filter using a 0.22-μm membrane filter (Thermo Scientific cat. no. 50-109-8762)
- Store up to 1 year at room temperature or 4°C
Elution buffer 1 (1 L)
- 20 ml 1 M MES·HCl, pH 6.5
- 5 ml 5 M imidazole, pH 8.0 (see recipe)
- 0.2 ml 1 M TCEP (see recipe)
- 480 g urea (Thermo Scientific cat. no. 036429.A4)
- 250 ml 1 M Arg/Glu, pH 6.5 (see recipe)
- Bring volume to 1 L with ddH2O or Milli-Q water.
- Filter using a 0.22-μm filter membrane.
- Store at 4°C
Composition: 20 mM MES·HCl, pH 6.5, 500 mM imidazole, 0.2 mM TCEP, 8 M urea, 250 mM Arg/Glu.
Elution buffer 2 (1 L)
- 100 ml 5 M imidazole, pH 8.0 (see recipe)
- 0.1 ml 1 M TCEP (see recipe)
- 3.28 g sodium acetate (Sigma-Aldrich EC No. 204-823-8)
- Fill to final volume with ddH2O or Milli-Q water.
- Filter using a 0.22-μm filter membrane.
- Store at 4°C
Composition: 500 mM imidazole, 0.1 mM TCEP, 40 mM sodium acetate, pH 5.
Elution buffer 3 (1 L)
- 60 ml 5 M imidazole (see recipe)
- 0.2 ml 1 M TCEP (see recipe)
- 20 ml 1 M Tris·Cl, pH 7.5 (see recipe)
- Fill to final volume with ddH2O or Milli-Q water.
- Filter using a 0.22-μm filter membrane.
- Store at 4°C
Composition: 20 mM Tris·Cl, pH 7.5, 300 mM imidazole, 0.2 mM TCEP.
Elution buffer 4 (1 L)
- 100 ml 5 M imidazole (see recipe)
- 1 ml 1 M TCEP (see recipe)
- 20 ml 1 M Tris·Cl, pH 7.5 (see recipe)
- Fill to final volume with ddH2O or Milli-Q water.
- Filter using a 0.22-μm filter membrane.
- Store at 4°C
- Just before use, crush one protease inhibitor table per 50 ml of loading buffer, add, and vortex to dissolve.
- Composition: 500 mM imidazole, 1 mM TCEP, 20 mM Tris·Cl pH 7.8, 1 protease inhibitor tablet.
Imidazole, 5 M, pH 8.0 (100 ml)
- 34.04 g imidazole (ACROS cat. no. 122020020)
- Dissolve in 70 ml of ddH2O or Milli-Q water
- Adjust pH to 8.0 with HCl
- Bring volume to 100 ml with ddH2O or Milli-Q water
- Sterilize by filtration using a 0.22-μm membrane filter (Thermo Scientific cat. no. 50-109-8762)
- Store at room temperature or 4°C
Kanamycin stock, 50 mg/ml, 1000× (10 ml)
- 0.5 g kanamycin (Fisher cat. no. 50-213-386)
- Dissolve in 10 ml Milli-Q water
- Sterilize using a syringe filter unit (Millipore Sigma cat. no. SLGV033RS)
- Store up to 12 months at 4°C
LB medium (1 L)
- 10 g NaCl (Fisher Scientific, cat. no. S271-10)
- 10 g tryptone (Fisher Scientific, cat. no. BP1421-2)
- 5 g yeast extract (Fisher Scientific, cat. no. BP9727-2)
- Bring volume to 1 L with ddH2O
- Autoclave
- Store at room temperature
Loading buffer, 1 L
- 20 ml 1 M Tris·Cl, pH 7.5 (see recipe)
- 400 ml 5 M NaCl (see recipe)
- 5 ml 5 M imidazole (see recipe)
- 0.5 ml 1 M TCEP (see recipe)
- Fill to final volume with ddH2O or Milli-Q water
- Filter using a 0.22-μm filter membrane
- Store at 4°C
- Just before use, crush one protease inhibitor table per 50 ml of loading buffer, add, and vortex to dissolve.
Composition: 20 mM Tris·Cl pH 7.5, 2 M NaCl, 25 mM imidazole, 0.5 mM TCEP, 1 protease inhibitor tablet.
Low-salt wash 1, 1 L
- 20 ml 1 M Tris·Cl, pH 7.5 (see recipe)
- 5 ml 5 M imidazole, pH 8.0 (see recipe)
- 0.2 ml 1 M TCEP (see recipe)
- 480 g urea (Thermo Scientific cat. no. 036429.A4)
- Bring volume to 1 L with ddH2O or Milli-Q water
- Filter using a 0.22-μm filter membrane
- Store at 4°C
Composition: 20 mM Tris·Cl, pH 7.5, 25 mM imidazole, 0.2 mM TCEP, 8 M urea.
Low-salt wash 2 (1 L)
- 20 ml 1 M Tris·Cl, pH 7.5 (see recipe)
- 5 ml 5 M imidazole, pH 8.0 (see recipe)
- 0.2 ml 1 M TCEP (see recipe)
- Fill to final volume with ddH2O or Milli-Q water
- Filter using a 0.22-μm filter membrane
- Store at 4°C
Composition: 20 mM Tris·Cl, pH 7.5, 25 mM imidazole, 0.2 mM TCEP.
Lysis buffer (1 L)
- 20 ml 1 M Tris·Cl, pH 7.5 (see recipe)
- 5 ml 5 M imidazole, pH 8.0 (see recipe)
- 0.1 ml 1 M TCEP (see recipe)
- 573 g guanidinium·HCl (Thermo Fisher Scientific cat. no. 24110)
- Bring volume to 1 L with ddH2O or Milli-Q water
- Filter using a 0.22-μm filter membrane
- Store at 4°C
Composition: 20 mM Tris·Cl, pH 7.5, 25 mM imidazole, 0.1 mM TCEP, 6 M guanidinium·HCl.
Magnesium chloride, 1 M (100 ml)
- 9.52 g magnesium chloride (Thermo Fisher Scientific cat. no. 223211000)
- Dissolve in 100 ml ddH2O or Milli-Q water
- Sterilize by filtration using a 0.22-μm membrane filter (Thermo Scientific cat. no. 50-109-8762)
- Store up to 2 months at −20°C
Magnesium chloride, 100 mM (1000 ml)
- 9.52 g magnesium chloride (Thermo Fisher Scientific cat. no. 223211000)
- Dissolve in 1000 ml ddH2O or Milli-Q water
- Sterilize by filtration using a 0.22-μm membrane filter (Thermo Scientific cat. no. 50-109-8762)
- Store up to 12 months at 4°C
NaCl, 5 M (1 L)
- 292.2 g NaCl (Fisher Scientific, cat. no. S271-10)
- Dissolve in 1 L of ddH2O or Milli-Q water
- Sterilize by filtration using a 0.22-μm membrane filter (Thermo Scientific cat. no. 50-109-8762)
- Store at room temperature or 4°C
NaN3, 20% (w/v) (50 ml)
- 10 g NaN3 (Fisher Scientific, cat. no. AAJ2161022)
- Dissolve in 50 ml of ddH2O or Milli-Q water
- Store at room temperature
Phosphorylation buffer (50 ml)
- 2.5 ml 1 M Tris·Cl, pH 7.5 (see recipe)
- 1.5 ml 5 M NaCl (see recipe)
- 0.01 ml 1 M TCEP (see recipe)
- Fill to final volume with ddH2O or Milli-Q water
- Filter using a 0.22-μm filter membrane
- Store at 4°C
Composition: 50 mM Tris·Cl pH 7.5, 150 mM NaCl, 0.2 mM TCEP.
SPA 1 (1 L)
- 20 ml 1 M MES·HCl, pH 6.0
- 240 g urea (Thermo Scientific cat. no. 036429.A4)
- 0.1 ml 1 M TCEP (see recipe)
- Bring volume to 1 L with ddH2O or Milli-Q water.
- Filter using a 0.22-μm filter membrane.
- Store at 4°C
Composition: 20 mM MES·HCl, pH 6.0, 4 M urea, 0.1 mM TCEP.
SPA 2 (1 L)
- 0.1 ml 1 M TCEP (see recipe)
- 1.64 g sodium acetate (Sigma-Aldrich EC No. 204-823-8)
- Fill to final volume with ddH2O or Milli-Q water
- Filter using a 0.22-μm filter membrane
- Store at 4°C
Composition: 20 mM sodium acetate·HCl, pH 5, 0.1 mM TCEP.
SPB 1 buffer (1 L)
- 20 ml 1 M MES·HCl, pH 6.0
- 240 g urea (Thermo Scientific cat. no. 036429.A4)
- 0.1 ml 1 M TCEP (see recipe)
- 400 ml 5 M NaCl (see recipe)
- Fill to final volume with ddH2O or Milli-Q water
- Filter using a 0.22-μm filter membrane
- Store at 4°C
Composition: 20 mM MES·HCl, pH 6.0, 4 M urea, 0.1 mM TCEP, 2 M NaCl.
SPB 2 buffer (1 L)
- 0.1 ml 1 M TCEP (see recipe)
- 1.64 g sodium acetate (Sigma-Aldrich EC no. 204-823-8)
- 400 ml 5 M NaCl (see recipe)
- Fill to final volume with ddH2O or Milli-Q water
- Filter using a 0.22-μm filter membrane
- Store at 4°C
Composition: 20 mM sodium acetate·HCl, pH 5, 0.1 mM TCEP, 2 M NaCl.
Spectinomycin stock solution, 50 mg/ml, 1000× (10 ml)
- 0.5 g spectinomycin (Fisher cat. no. BP29571)
- Dissolve in 10 ml Milli-Q water
- Sterilize using a syringe filter unit (Millipore Sigma cat no. SLGV033RS)
- Store up to 6 months at −20°C
Storage buffer (50 ml)
- 40 ml 1 M Arg/Glu, pH 7.5
- 2.5 ml 1 M Tris·Cl, pH 7.5 (see recipe)
- 1.5 ml 5 M NaCl (see recipe)
- 0.01 ml 1 M TCEP (see recipe)
- Fill to final volume with ddH2O or Milli-Q water.
- Filter using a 0.22-μm filter membrane.
- Store at 4°C
Composition: 50 mM Tris·Cl, pH 7.5, 800 mM Arg/Glu, 150 mM NaCl, 0.2 mM TCEP.
TB medium (1 L)
- 12 g tryptone (Fisher Scientific cat. no. BP1421-2)
- 24 g yeast extract (Fisher Scientific cat. no. BP9727-2)
- 4 g glycerol (EC No. 200-289-5)
- 2.3 g KH2PO4 (Sigma-Aldrich EC no. 231-913-4)
- 16.4 g K2HPO4 (Sigma-Aldrich EC no. 231-834-5)
- Bring volume to 1 L with ddH2O
- Autoclave
- Store at room temperature
TCEP, 1 M (10 ml)
- 2.5 g tris(2-carboxyethyl)phosphine (TCEP; Biosynth cat. no. FT01756)
- Dissolve in 10 ml ddH2O or Milli-Q water
- Sterilize by filtration using a 0.22-μm syringe filter (Thermo Scientific cat. no. 50-109-8762)
- Aliquot into 1-ml fractions
- Store up to 2 months at −20°C
Tris·Cl, 1 M, pH 7.5 (1 L)
- 121.14 g Tris·Cl (Fisher cat. no. BP152-1)
- Dissolve in 800 ml ddH2O or Milli-Q water
- Adjust pH to 7.5 with HCl, then add H2O or Milli-Q water to a final volume of 1 L
- Sterilize by filtration using a 0.22-μm membrane filter (Thermo Scientific cat. no. 50-109-8762)
- Store at room temperature or 4°C
Urea, 8 M (100 ml)
- 48.48 g urea (Thermo Scientific cat. no. 036429.A4)
- Dissolve in 100 ml ddH2O or Milli-Q water (heat can increase solubility, but do not exceed 37°C)
- Sterilize by filtration using a 0.22-μm membrane filter (Thermo Scientific cat. no. 50-109-8762)
- Store up to 12 months at room temperature
COMMENTARY
Background Information
In this article, we delineate procedures for expressing, purifying, and ensuring solubility of U1-70K and its BAD1 and BAD2 domains in various phosphorylated states. Both unphosphorylated U1-70K and its BAD domains can be purified similarly. The typical yield of U1-70K is ∼0.4 mg of protein per liter of TB. However, the protein yield may vary based on factors including the quality of the transformed cells, the type of medium employed, the speed of protein purification, and the condition of the cells. Therefore, it is advisable to use freshly transformed cells. Additionally, we recommend expediting the protein purification process insofar as possible without compromising its integrity. Besides these suggestions, the following parameters deserve special consideration in the purification of U1-70K and BAD domains.
Critical Parameters
Our recent study revealed that U1-70K undergoes phase separation in a physiological buffer. We discovered that the interaction between the BAD regions and the RRM are responsible for this phase separation. Phase separation poses significant challenges for the biophysical characterization of proteins. Protein droplets, being denser than buffers, cause phase-separating proteins to precipitate during centrifugation and incubation, processes commonly used in protein purification and concentration. Although phase-separated droplets can be redissolved, some proteins in droplets age and harden over time, eventually transforming into irreversible aggregates.
Purification of U1-70K
The most critical parameter for the purification of U1-70K is the concentration of Arg/Glu. U1-70K is soluble at high concentrations of Arg/Glu; however, arginine concentrations of >100 mM are incompatible with the Ni-NTA resin. To circumvent this problem, we utilize guanidinium·HCl and urea, as arginine can elute His-tagged proteins. Unlike arginine, guanidinium·HCl and urea are compatible with Ni-NTA resin. Additionally, high concentrations of arginine or guanidinium HCl can interfere with protein binding to ion-exchange columns. Although urea is compatible with ion-exchange chromatography, it can cause carbamylation of proteins after a long incubation time (Sun et al., 2014). Therefore, it is important to exchange the U1-70K protein into a buffer without urea as quickly as possible.
Expression of U1-70K
To optimize the expression of U1-70K, we co-transform E. coli with the pBB535 plasmid, which encodes DnaK and DnaJ. Based on our experience, these chaperones are necessary to express full-length U1-70K. It is also necessary to use BL21 Star cells transformed with the pRARE plasmid from Rosetta cells. Using other cells results in a major truncated species that is difficult to separate from full-length U1-70K. We have only had success expressing the full-length U1-70K using a codon-optimized sequence in the pSMT3 vector.
Phosphorylation of U1-70K
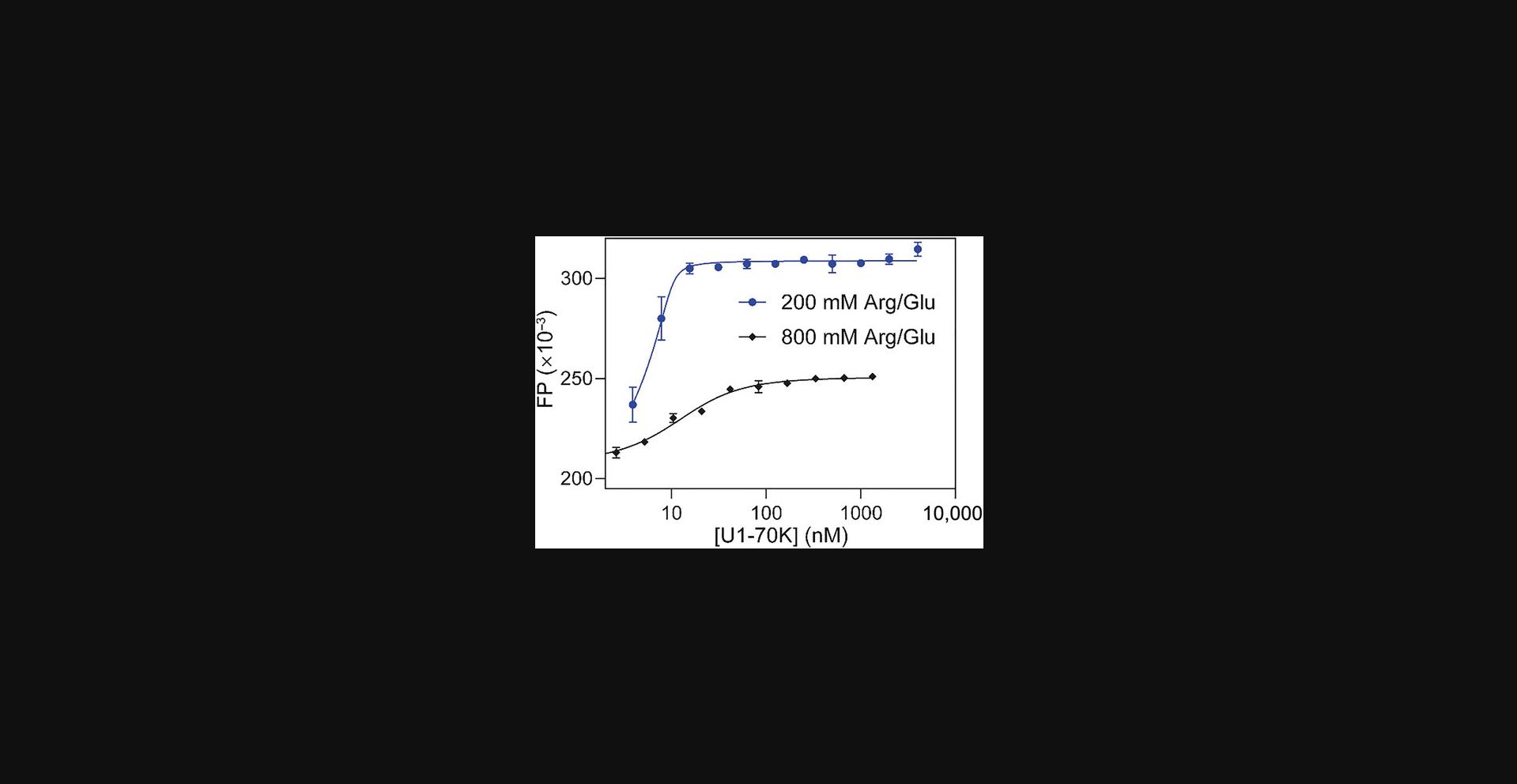
The most critical parameter for the U1-70K phosphorylation is the arginine concentration in the buffer. In our experience, an arginine concentration in the final phosphorylation buffer of >100 mM inhibits phosphorylation. However, unphosphorylated U1-70K is insoluble in buffers with <100 mM arginine. Therefore, dialysis of U1-70K initially in 800 mM arginine into 100 mM arginine in the presence of SRPK1 is the optimal approach.
Purification of U1-70K BAD1
The most critical parameter for the U1-70K BAD1 purification is the use of BL21-CodonPlus(DE3) cells. Utilizing cells that did not have the pRARE plasmid resulted in arginine residues wobbling to lysine residues. Using the pBB535 plasmid reduces the production of truncated products, which is important because these are difficult to separate from full-length BAD1. In our experience, BAD1 is insoluble in the absence of 6 M guanidinium·HCl until after the Ni-NTA purification step. We speculate that this is due to nonspecific interactions with nucleic acids. We have only had success in expressing the BAD1 domain using a codon-optimized sequence in the pSMT3 vector. We have added a tryptophan residue to the C-terminus of the BAD1 domain. This serves two useful functions: first, it facilitates quantification of BAD1; second, it makes it possible to confirm the presence of full-length BAD1 by collecting fluorescence spectra.
Phosphorylation of BAD1
The most critical parameter for BAD1 phosphorylation is that BAD1 becomes less soluble after phosphorylation.
Purification of BAD2
The most critical parameter for the BAD2 purification is the use of the pSMT3 vector. This is because the domain is too short to be expressed without the SUMO tag.
Troubleshooting
Table 1 delineates possible problems that may be encountered in performing Basic Protocols 1-5 as well as solutions to those problems.
| Problem | Possible cause | Solution |
|---|---|---|
| Protein degradation after purification (U1-70K) | Microbes grew in the storage buffer used to store the protein, degrading the sample. | Purify again and make a new storage buffer. |
| Protein degradation after SP column (U1-70K) | An incorrect cell strain was used, and the sample includes one or more truncated species that are almost impossible to separate from the full-length protein. | The truncated species are very difficult to separate from the full-length protein. Use BL21 Star pRARE/pBB535 cells to avoid obtaining these species. |
| Protein is insoluble (U1-70K) | A buffer with <800 mM Arg/Glu was used. | Solubilize in buffer using 8 M urea, and then buffer exchange into a buffer containing 800 mM Arg/Glu. U1-70K is poorly soluble in 400 mM Arg/Glu. |
| Contamination bands are observed | The Ni-NTA, ion exchange column, or SEC column is dirty. | Clean all columns according to the protocol above or the user manuals. |
| The washing step in the Ni-NTA step was too fast. | The flow rate in the Ni-NTA washing step should be <10 ml/min. | |
| The gradient of the ion-exchange step was too steep. | Be sure to perform the FPLC step exactly as described in the relevant protocol. | |
| Ulp1 cleavage was incomplete. | Allow sufficient cleavage time, as described in the protocol. The cleavage will take ∼1 hr to finish at 37°C, or longer at lower temperatures. | |
| Protein loss during concentration | The molecular weight cutoff (MWCO) of the concentrator is too large. | In general, the MWCO of the concentrator should be less than half the protein MW (however, a different cutoff applies to U1-70K; see below). |
| Membrane leakage has occurred | Use the correct centrifugal speed recommended by the manual to avoid damaging the membrane. | |
| Protein phase separation or aggregation can also cause apparent protein loss. Aggregation will be evidenced by white flakes in your protein sample; phase separation may be impossible to see during this step. |
Ensure correct concentrations of Arg/Glu are used to solubilize proteins. The sample should be inverted to mix after every 15 min of centrifugation. Avoid centrifugation for >15 min. |
|
| No protein expression is seen | The pSMT3 vector was not used to express the protein. We have not had any success using vectors other than pSMT3 to express any U1-70K protein constructs or the full-length protein. | Use the pSMT3 expression vector. |
| SUMO-U1-70K is in the flowthrough of the spin concentrator | The filter membrane MWCO is too large or the filter is leaking. U1-70K flows through a concentrator with a MWCO of ≥30 kDa. | Use a 10-kDa-MWCO spin concentrator. |
Time Considerations
Basic Protocols 1, 3, and 5
The timeframes for the purification of U1-70K, BAD1, and BAD2 are essentially the same. Rough time estimate are provided in the protocols. More detailed time considerations are as follows.
- Preparation of media and buffers: 2-3 hr (if common stock solutions are available in a biochemistry lab; if not, preparing all buffers may take 1-2 days)
- Transformation of competent cells: 1.5 hr plus overnight incubation to allow cell growth on plate
- Starter culture: 2 hr
- Growing cells: 6-8 hr
- Cell harvest: 4-5 hr, including collecting cells into centrifugal bottles (∼15 min), centrifugation (∼20 min), collecting cell pellets (∼10 min), and freeze/thaw cycles (3-4 hr)
- Purification of the protein: 12 hr total (thawing and lysing cells: 1 hr; separating soluble fractions: 1 hr; Ni-NTA column step: 2-3 hr; ion-exchange chromatography: 3-4 hr; buffer exchange out of urea: 3 hr)
Basic Protocol 2
The phosphorylation of SUMO-U1-70K using SRPK1 takes ∼1 day 8 hr:
- Phosphorylation of the protein: 1 day
- Running the ion-exchange column: 2-3 hr
- Running the SDS-PAGE gel: 2 hr
- Concentration and buffer exchange of samples containing phosphorylated protein: 2 hr
Basic Protocol 4
The phosphorylation of the BAD1 domain using SRPK1 takes ∼8 hr:
- Phosphorylation of the protein: 1 hr
- Running the ion-exchange column: 2-3 hr
- Running the SDS-PAGE gel: 2 hr
- Concentration and buffer exchange of samples containing phosphorylated protein: 2 hr
Support Protocol 1
- Transformation of competent cells: 1.5 hr plus overnight incubation
- Preparation and sterilization of medium and buffers: 2 hr
- Sterilization of solids: 2 hr
- Starter culture: 2 hr
- Growing cells: 4 hr
- Making cells chemically competent: 4 hr
Support Protocol 2
The timeframes for purification of SUMO-SRPK1 are as follows:
- Preparation of media and buffers: 2-3 hr (if common stock solutions are available in a biochemistry lab; if not, preparing all buffers may take 1-2 days)
- Transformation of competent cells: 1.5 hr plus overnight incubation
- Starter culture: 2 hr
- Growing cells: 6-8 hr
- Cell harvest: 4-5 hr, including collecting cells into centrifugal bottles (∼15 min), centrifugation (∼20 min), collecting cell pellets (∼10 min), and freeze/thaw cycles (3-4 hr)
- Purification of the protein: 8 hr total (thawing and lysing cells: 1 hr; separating soluble fractions: 1 hr; Ni-NTA column step: 2-3 hr; ion-exchange chromatography step: 3-4 hr; concentrating protein: ∼1 hr (8 hr total)
Acknowledgments
This work is supported by the U.S. National Institutes of Health (R35GM147091-01 to Dr. Jun Zhang).
Author Contributions
Trent Paul : Data curation; investigation; methodology; visualization; writing—original draft. Shariq Jamal : Investigation. Ethan Ekpenyong : Investigation. Peter Prevelige : Investigation. Talia Fargason : Investigation; writing—review and editing. Zihan Zhang : Investigation. Jun Zhang : Conceptualization; data curation; formal analysis; funding acquisition; investigation; methodology; project administration; supervision; validation; writing—review and editing.
Conflict of Interest
The authors declare no conflict of interest.
Open Research
Data Availability Statement
All data are included in this protocol.
Literature Cited
- Bishof, I., Dammer, E. B., Duong, D. M., Kundinger, S. R., Gearing, M., Lah, J. J., Levey, A. I., & Seyfried, N. T. (2018). RNA-binding proteins with basic-acidic dipeptide (BAD) domains self-assemble and aggregate in Alzheimer's disease. Journal of Biological Chemistry , 293(28), 11047–11066. https://doi.org/10.1074/jbc.RA118.001747
- Cao, W., & Garcia-Blanco, M. A. (1998). A serine/arginine-rich domain in the human U1 70k protein is necessary and sufficient for ASF/SF2 binding. Journal of Biological Chemistry , 273(32), 20629–20635. https://doi.org/10.1074/jbc.273.32.20629
- Cho, S., Hoang, A., Sinha, R., Zhong, X. Y., Fu, X. D., Krainer, A. R., & Ghosh, G. (2011). Interaction between the RNA binding domains of Ser-Arg splicing factor 1 and U1-70K snRNP protein determines early spliceosome assembly. Proceedings of the National Academy of Sciences USA , 108(20), 8233–8238. https://doi.org/10.1073/pnas.1017700108
- Diner, I., Hales, C. M., Bishof, I., Rabenold, L., Duong, D. M., Yi, H., Laur, O., Gearing, M., Troncoso, J., Thambisetty, M., Lah, J. J., Levey, A. I., & Seyfried, N. T. (2014). Aggregation properties of the small nuclear ribonucleoprotein U1-70K in Alzheimer disease. Journal of Biological Chemistry , 289(51), 35296–35313. https://doi.org/10.1074/jbc.M114.562959
- Gallagher, S. (2006). One-dimensional SDS gel electrophoresis of proteins. Current Protocols in Molecular Biology , 75, 10.2A.1–10.2A.37. https://doi.org/10.1002/0471142727.mb1002as75
- Hu, Z., Li, M., Huo, Z., Chen, L., Liu, S., Deng, K., Lu, X., Chen, S., Fu, Y., & Xu, A. (2022). U1 snRNP proteins promote proximal alternative polyadenylation sites by directly interacting with 3′ end processing core factors. Journal of Molecular Cell Biology , 14(8), mjac054. https://doi.org/10.1093/jmcb/mjac054
- Kattah, N. H., Kattah, M. G., & Utz, P. J. (2010). The U1-snRNP complex: Structural properties relating to autoimmune pathogenesis in rheumatic diseases. Immunological Reviews , 233(1), 126–145. https://doi.org/10.1111/j.0105-2896.2009.00863.x
- Kondo, Y., Oubridge, C., van Roon, A. M., & Nagai, K. (2015). Crystal structure of human U1 snRNP, a small nuclear ribonucleoprotein particle, reveals the mechanism of 5′ splice site recognition. Elife , 4, e04986. https://doi.org/10.7554/eLife.04986
- Lee, E. S., Smith, H. W., Wolf, E. J., Guvenek, A., Wang, Y. E., Emili, A., Tian, B., & Palazzo, A. F. (2022). ZFC3H1 and U1-70K promote the nuclear retention of mRNAs with 5′ splice site motifs within nuclear speckles. RNA , 28(6), 878–894. https://doi.org/10.1261/rna.079104.122
- Moore, M. J., Query, C. C., & Sharp, P. A. (1993). Splicing of precursors to mRNA by the spliceosome. In R. F. Gesteland & J. F. Atkins (Eds.), The RNA world (pp. 303–357). Cold Spring Harbor Laboratory Press.
- Northemann, W., Berg, H., Stahnke, G., Walter, M., Hunt, N., & Fenning, S. (1995). Identification of an inhibitory element within the human 68-kDa (U1) ribonucleoprotein antigen. Protein Expression and Purification , 6(6), 748–756. https://doi.org/10.1006/prep.1995.0005
- Richter, C., Simon, T., Asen, I., Brenner-Weiss, G., & Hubbuch, J. (2014). Soluble full-length expression and characterization of snRNP protein U1-68/70K. Protein Expression and Purification , 104, 65–70. https://doi.org/10.1016/j.pep.2014.08.009
- Sun, S., Zhou, J. Y., Yang, W., & Zhang, H. (2014). Inhibition of protein carbamylation in urea solution using ammonium-containing buffers. Analytical Biochemistry , 446, 76–81. https://doi.org/10.1016/j.ab.2013.10.024
- Tazi, J., Kornstädt, U., Rossi, F., Jeanteur, P., Cathala, G., Brunel, C., & Lührmann, R. (1993). Thiophosphorylation of U1-70K protein inhibits pre-mRNA splicing. Nature , 363(6426), 283–286. https://doi.org/10.1038/363283a0
- Woppmann, A., Patschinsky, T., Bringmann, P., Godt, F., & Luhrmann, R. (1990). Characterisation of human and murine snRNP proteins by two-dimensional gel electrophoresis and phosphopeptide analysis of U1-specific 70K protein variants. Nucleic Acids Research , 18(15), 4427–4438. https://doi.org/10.1093/nar/18.15.4427
- Xue, S., Gong, R., He, F., Li, Y., Wang, Y., Tan, T., & Luo, S. Z. (2019). Low-complexity domain of U1-70K modulates phase separation and aggregation through distinctive basic-acidic motifs. Science Advances , 5(11), eaax5349. https://doi.org/10.1126/sciadv.aax5349
- Zhang, S., Aibara, S., Vos, S. M., Agafonov, D. E., Lührmann, R., & Cramer, P. (2020). Structure of a transcribing RNA polymerase II–U1 snRNP complex. bioRxiv, https://doi.org/10.1101/2020.10.19.344200 bioRxiv
- Zillmann, M., Rose, S. D., & Berget, S. M. (1987). U1 small nuclear ribonucleoproteins are required early during spliceosome assembly. Molecular and Cellular Biology , 7(8), 2877–2883. https://doi.org/10.1128/mcb.7.8.2877-2883.1987

