Expanded Antigen-Specific Elimination Assay to Measure Human CD8+ T Cell Cytolytic Potential
David R. Collins, David R. Collins, Mpho J. Olatotse, Mpho J. Olatotse, Zachary J. Racenet, Zachary J. Racenet, Umar Arshad, Umar Arshad, Elif Çakan, Elif Çakan, Gaurav D. Gaiha, Gaurav D. Gaiha, Kiera L. Clayton, Kiera L. Clayton, Bruce D. Walker, Bruce D. Walker
Abstract
Durable cellular immunity against pathogens is dependent upon a coordinated recall response to antigen by memory CD8+ T cells, involving their proliferation and the generation of secondary cytotoxic effector cells. Conventional assays measuring ex vivo cytotoxicity fail to capture this secondary cytolytic potential, especially in settings where cells have not been recently exposed to their cognate antigen in vivo. Here we describe the expanded antigen-specific elimination assay (EASEA), a flow cytometric endpoint assay to measure the capacity of human CD8+ T cells to expand in vitro upon antigen re-exposure and generate secondary effector cells capable of selectively eliminating autologous antigen-pulsed target cells across a range of effector-to-target ratios. Unlike alternative assays, EASEA avoids the hazards of radioactive labeling and viral infection and can be used to study responses to individual or pooled antigens of interest. © 2024 The Author(s). Current Protocols published by Wiley Periodicals LLC.
Basic Protocol : Expanded antigen-specific elimination assay
INTRODUCTION
Adaptive cellular immunity against intracellular pathogens and tumors is mediated by cytotoxic CD8+ T cells, whose T cell receptors (TCRs) recognize antigenic peptides presented by class I major histocompatibility complex proteins on the surface of infected cells. Such CD8+ T cells exercise effector function through their release of granules containing the cytotoxic effector molecules perforin and granzyme B to cause apoptosis of the target cells (reviewed in Russell & Ley, 2002). Upon antigen clearance, CD8+ T cells persist in a functional but quiescent memory state characterized by low expression of cytotoxic effectors and high proliferative potential. These functional cells respond to antigen re-exposure via programmed T cell differentiation pathways that result in the generation of secondary effector cells with high cytolytic capacity (Migueles et al., 2020). However, under some conditions, some CD8+ T cells persist in a dysfunctional state characterized by markedly reduced proliferative potential and diminished expression of cytotoxic effector molecules (reviewed in Wherry, 2011). Such dysfunctional quiescent CD8+ T cells may be indistinguishable from functional quiescent CD8+ T cells by conventional assays usually used to measure ex vivo cytotoxicity. This is especially true for cells collected from tissues in which the expression of cytolytic effector proteins is further suppressed under homeostatic conditions marked by the absence of active pathogen replication and inflammation (Collins et al., 2023; Reuter et al., 2017). However, in vitro re-exposure to antigen, such as that employed in the assay described here, enables robust assessment of the secondary cytotoxic potential of memory CD8+ T cells and can distinguish functional from dysfunctional cells irrespective of recent in vivo antigen exposure.
Here we describe the expanded antigen-specific elimination assay (EASEA), a flow cytometric method to quantitatively measure the ability of human antigen-specific memory CD8+ T cell responses to eliminate autologous peptide-pulsed target cells during a 4-hr co-incubation after 6 days of peptide-specific effector cell expansion. EASEA provides numerous advantages relative to alternative cytotoxicity assays such as chromium release (Brunner et al., 1968), target cell apoptotic activity (Fischer et al., 2002; Jerome et al., 2003; Liu et al., 2002), and virus inhibition assays (Fauce et al., 2007; Saez-Cirion et al., 2010; Yang et al., 1997), avoiding the hazards of radiolabeling and virus infection while enabling measurement of the cytolytic potential of antigen-specific memory CD8+ T cell responses against specific antigens presented on autologous target cells. In this protocol we describe the assay using HIV-epitope-specific CD8+ T cell effectors derived from peripheral blood and autologous CD4+ T cell targets pulsed with cognate peptide. However, EASEA is adaptable for use with various target cell types pulsed with individual or pooled peptide antigens, as well as with primary human cells derived from different tissue origins.
CAUTION : Primary human cells should be obtained only after informed consent under protocols approved by institutional review for human subjects research and should be handled under appropriate Biosafety Level 2 (BSL-2) or higher sterile conditions.
STRATEGIC PLANNING
As EASEA is not suitable for high-throughput response screening because of its relatively cell-intensive requirements, individual antigen-specific CD8+ T cell responses of interest should be identified beforehand using screening assays such as the immunospot assay. Alternatively, pooled peptides covering a protein or set of proteins of interest may be used.
NOTE : Appropriate informed consent is necessary for obtaining and use of human study material.
Basic Protocol: EXPANDED ANTIGEN-SPECIFIC ELIMINATION ASSAY
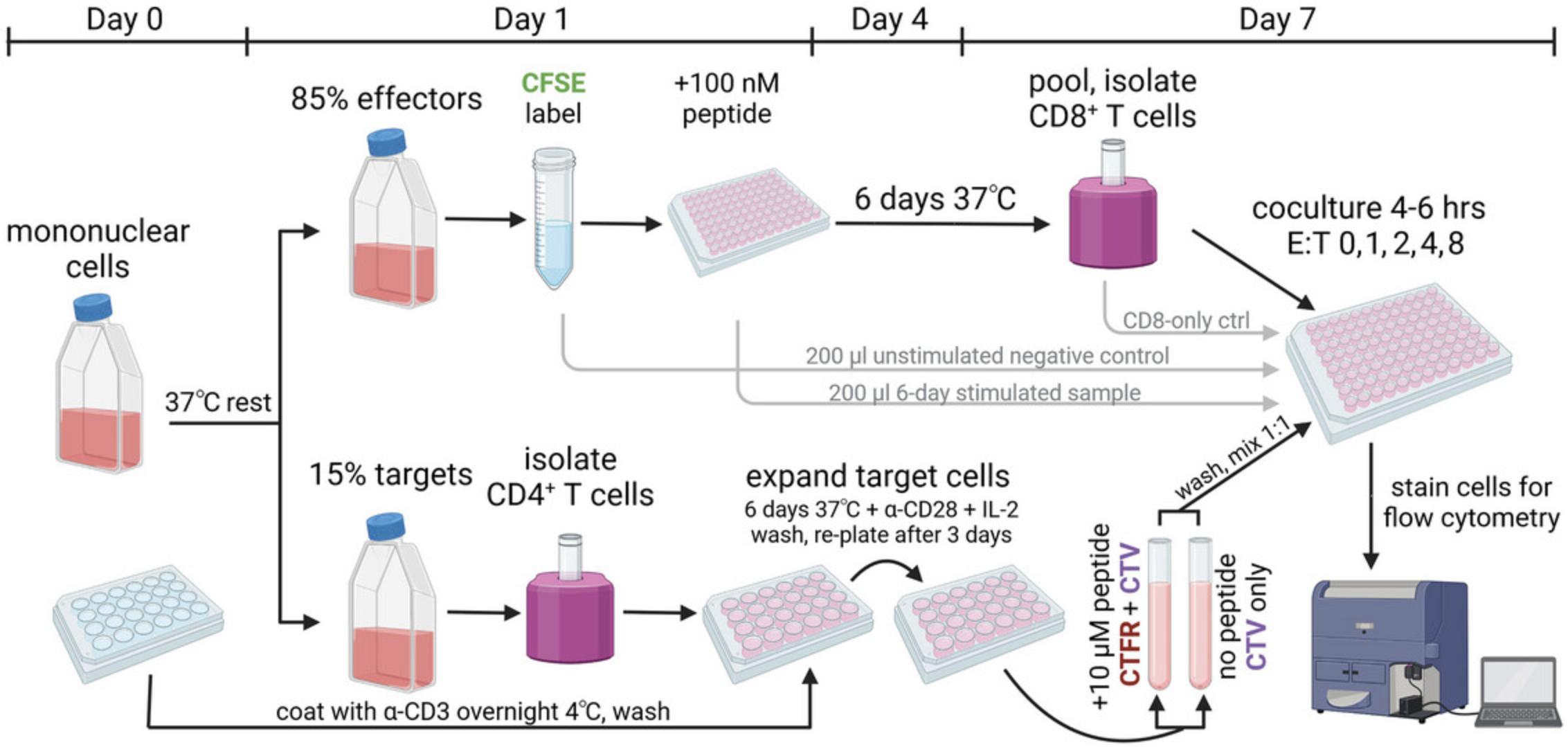
This demonstration protocol focuses on the assay of cytotoxic T cells developing in people with HIV infection and thus harboring HIV antigen-specific CD8+ T cells. It begins with peripheral blood or tissue-derived mononuclear cells isolated from research participants, which are then split into two pools for subsequent culture (Fig. 1). The first pool of cells, the effector cell pool, is stimulated with antigenic peptide(s) of interest in the absence of exogenous cytokines; this allows antigen-specific CD8+ T cell (effector cell) proliferation that can be subsequently tracked by dilution of carboxyfluorescein (CFSE). The second pool of cells, the target cell pool, is nonspecifically expanded via TCR stimulation after isolation of target cells. 50% of these cells are loaded with the cognate peptide(s) of interest and labeled with a unique fluorescent dye for tracking of their selective elimination across varying effector:target (E:T) ratios during a 4-hr coculture. When implemented successfully, the assay measures titratable reductions in residual peptide-pulsed target cells by secondary effector CD8+ T cells at increasing E:T ratios, thus enabling the quantitation and comparison of antigen-specific cytolytic potential via area-under-curve (AUC) analyses.
Materials
-
Ultra-LEAF purified anti-CD3 antibody (clone OKT3; Biolegend, cat. no. 317304)
-
Carbonate coating buffer (Thermo, cat. no. CB01100, or alternatively see recipe)
-
R10 culture medium (see recipe)
-
Micrococcal nuclease S7 (Sigma, cat. no. 10107921001)
-
EasySep CD4+ T cell isolation kit (StemCell Technologies, cat. no. 17952)
-
PBS (Corning, cat. no. 21040CM)
-
Recombinant human IL-2 (R&D, cat. no. 020-IL-010)
-
Ultra-LEAF purified anti-CD28 antibody (clone CD28.2; Biolegend, cat. no. 302902)
-
CellTrace CFSE (Thermo, cat. no. C34554)
-
Dimethyl sulfoxide (DMSO)
-
Peptide(s) of interest: 200 µM working stocks diluted in RPMI (custom-synthesized or commercially purchased)
-
TC-treated polystyrene 24-well plates (Corning, cat. no. 3524)
-
EasySep CD8+ T cell isolation kit (StemCell Technologies, cat. no. 17953)
-
CellTrace FarRed (Thermo, cat. no. C34564)
-
CellTrace Violet (Thermo, cat. no. C34557)
-
FACS buffer (see recipe)
-
Optional : APC-conjugated pHLA multimer(s) of interest (custom-synthesized or commercially purchased)
-
BV605-conjugated anti-CD3 (clone SK7; Biolegend, cat. no. 344836)
-
BV711-conjugated anti-CD4 (clone RPA-T4; Biolegend, cat. no. 300557)
-
BUV395-conjugated anti-CD8 (clone RPA-T8; BD Biosciences, cat. no. 563795)
-
LiveDead Near-IR fixable viability dye (Thermo, cat. no. L34975)
-
Cytofix Fixation Solution (BD Biosciences cat. no. 554655, or 4% paraformaldehyde)
-
Biosafety Level 2 cabinet (BL2+ if protocol is adapted for use of target cells infected by HIV-1)
-
Untreated polystyrene 24-well plates (Corning, cat. no. 3738)
-
Plate seals (MP Biomedicals, cat. no. 0976401C2)
-
4°C refrigerator
-
25-cm2 (T25) cell culture flasks (Corning, cat. no. 430639)
-
Tabletop centrifuge with conical tube and plate rotors
-
37°C, 5% CO2 incubator
-
EasySep magnets (StemCell cat. no. 18000)
-
Microscope and hemacytometer or automated cell counter
-
5-ml polystyrene tubes (Corning, cat. no. 352052)
-
15- and 50-ml polypropylene conical tubes
-
96-well polystyrene round-bottom plates (Corning, cat. no. 3879)
-
200-µl multichannel pipette
-
Sterile reagent reservoirs (Argos Technologies, cat. no. B3110-50, or similar)
-
Flow cytometer (BD Fortessa or Symphony 5-laser with FACSDiva software, or similar)
-
FlowJo analysis software (BD Biosciences)
Day 0 (1-1.5 hr)
1.Prepare plate for target cell expansion: Coat wells of an untreated 24-well plate with 400 µl/well of 2 µg/ml anti-CD3 in carbonate coating buffer. Seal the plate and incubate overnight at 4°C.
2.Isolate mononuclear cells from peripheral blood or tissue specimens obtained from participants of interest after informed consent under Institutional Review Board (IRB)-approved protocols. If using cryopreserved specimens, thaw in R10 medium using established cell thawing protocols. We recommend adding thawed cells to 10 ml R10 with 10 U/ml micrococcal nuclease, centrifuging for 3 min at 800 RCF, room temperature, and then resuspending in R10 at ∼1-2 million cells/ml in an upright 25-ml (T25) flask for an overnight rest at 37°C, 5% CO2 to allow optimal cell recovery. Count viable cells using a microscope and hemacytometer or automated cell counter.
Day 1 (1.5-2.5 hr)
3.Establish separate cell pools (derived from mononuclear cells prepared above) for subsequent effector and target cell cultures: Count cells. For each response of interest, use 85% of cells (or ∼4-30 million cells) for peptide stimulation of effector cells and 15% of cells (or ∼1-6 million cells) for nonspecific target cell expansion.
4.Isolate target cells: Isolate CD4+ T cells from the designated target cell pool using 5-ml polystyrene tubes and EasySep CD4+ T cell isolation magnetic negative selection kit as per manufacturer's instructions. Count cells.
5.Expand isolated target cells: Remove seal from anti-CD3-coated 24-well plate. Remove anti-CD3 solution by aspiration and gently wash plate twice with 1 ml/well PBS to remove residual carbonate coating buffer.
6.Add 400 µl of pre-warmed R10 + 10 ng/ml (∼100 U/ml) IL-2 to the washed wells.
7.Centrifuge isolated CD4+ T cells for 3 min at 800 RCF, room temperature. Resuspend at a concentration of up to 2 million cells/ml in R10 + 10 ng/ml IL-2 and add 4 µg/ml of anti-CD28 antibody. Add cells in an additional 400 µl to each well of the 24-well plate (final volume 800 µl). Incubate at 37°C, 5% CO2 for 3 days.
8.Label effector cells in the designated effector cell pool: First, prepare 5× CFSE solution by thawing one vial of lyophilized CFSE, resuspending the contents in 18 µl DMSO, vortexing the mixture, and adding to 20 ml PBS. Second, centrifuge cells in the effector cell pool for 3 min at 800 RCF, room temperature, in a conical tube and resuspend in 800 µl PBS. Add 200 µl of 5× CFSE solution, mix, and incubate at 37°C, 5% CO2 for 20 min. After incubation, quench any remaining dye in solution by adding 10 ml of R10 medium, pre-warmed to 37°C.
9.Expand antigen-specific effector cells: Centrifuge CFSE-stained mononuclear cells in the effector cell pool for 3 min at 800 RCF, room temperature, and then resuspend in R10 medium at 1 million cells/ml. Split off 200 µl into one well of a 96-well round-bottom plate as an unstimulated negative control. Add HLA-optimal peptide(s) of interest to a final concentration of 100 nM to expand antigen-specific CD8+ T cells in the remaining mononuclear effector cell pool, and then plate 200 µl/well (200,000 cells/well) of the cell suspension into the 96-well plate(s). Incubate at 37°C, 5% CO2 for 6 day.
Day 3 (0.5-1 hr)
10.Expand target cells: After 3 days of stimulation, remove CD4+ T cells from coated wells, count cells, centrifuge 3 min at 800 RCF, room temperature, and resuspend cells in pre-warmed R10 + 10 ng/ml IL-2 at ∼1 million cells/ml. Plate 1 ml/well into a new TC-treated 24-well plate. Incubate at 37°C, 5% CO2 for 3 days.
Day 6 (6-8 hr; hands-on: 2-4 hr)
11.Peptide-pulse target cells: After an additional 3 days of expansion after the stimulation of CD4+ T cell targets, resuspend, pool, and count cells. Split cells into two conical tubes; leave one tube un-pulsed with peptide, and add 10 µM peptide(s) of interest to the second tube. Incubate at 37°C for 25 min.
12.Isolate effector cells: After 6 days of antigen-specific expansion, pool all wells of mononuclear cell populations containing CD8+ T cells expanded by each peptide by resuspending well contents using a multichannel pipettor and transferring into reagent reservoirs and then into a conical tube. After pooling, save 200 µl of unstimulated and peptide-stimulated mononuclear cells in one well of a 96-well round-bottom plate at 37°C for later assessment of antigen-specific expansion. While target cells are incubating with peptide(s), count and isolate CD8+ T cells from the remaining CFSE-stained, peptide-expanded mononuclear cell populations using 5-ml polystyrene tubes and an EasySep CD8+ T cell isolation magnetic negative selection kit as per the manufacturer's protocol. Count isolated CD8+ T cells.
13.Label target cells: Prepare stocks of CellTrace Far Red (CTFR) and CellTrace Violet (CTV) dyes by resuspending in 50 µl DMSO per vial. After 25 min of incubation with/without peptide(s) as in step 11, add 1 µl CTV per ml to both unpulsed and peptide-pulsed cells in 1 ml R10 medium, and add 1 µl CTFR per ml to only the peptide-pulsed cells in 1 ml pre-warmed R10.Incubate at 37°C, 5% CO2 for 5 min and then wash twice with 10 ml R10 medium. Resuspend final pellets in 1 ml R10 each and combine pulsed and unpulsed cells at a 1:1 ratio.
14.Coculture effector and target cells: Prepare a 96-well round-bottom plate for coculture across varying E:T ratios. A range of 25,000-150,000 target cells/well is recommended, depending upon effector and target cell yields. Assuming sufficient cells are available to use 50,000 target cells/well, dilute mixed target cells to a concentration of 500,000 cells/ml in R10 medium and plate 100 µl per well; centrifuge and resuspend effector cells at 4,000,000 cells/ml in R10 and plate 12.5 µl for E:T 1:1, 25 µl for E:T 2:1, 50 µl for E:T 4:1, and 100 µl for E:T 8:1.Add another 12.5 µl to an additional well as an effector-only control. Add R10 medium to bring all wells to 200 µl final volume and incubate at 37°C, 5% CO2 for 4-6 hr to allow cytotoxic elimination of target cells by effector cells.
15.Flow cytometric staining: After coculture, centrifuge plate for 3 min at 800 RCF, room temperature. Flick plate (or multichannel aspirate, if preferred) to discard supernatant into the appropriate biohazard waste container inside the biosafety cabinet. Resuspend cells in FACS buffer. Optionally, before surface staining, add 2-4 nM APC-conjugated peptide-HLA (pHLA) multimer(s) of interest to the effector-only and/or CFSE proliferation samples for 15 min at 4°C to assess antigen-specific TCR surface expression and frequency of antigen-specific effector cells. Add a cocktail of BV605-conjugated anti-CD3, BV711-conjugated anti-CD4, BUV395-conjugated anti-CD8 (each at 1:100 final dilution), and Live-Dead Near-IR (resuspended in 50 µl DMSO; unused portion can be refrozen at –20°C for future use) at 1:1000 final dilution. Incubate at 4°C for 30 m in and then wash with 200 µl/well FACS buffer. Fix cells with Cytofix fixation buffer or 4% paraformaldehyde for 20 min at 4°C, and then wash with 200 µl/well FACS buffer.
16.Compensate fluorescence overlap and acquire data using a flow cytometer, and then analyze data using flow cytometry analysis software as discussed below in the Understanding Results section.
REAGENTS AND SOLUTIONS
Carbonate coating buffer
- 8.4 g NaHCO3 (Sigma, cat. no. S6014)
- 3.56 g Na2CO3 (Sigma, cat. no. 223530)
- 1 L distilled, deionized water
- Adjust pH to 9.5
- Filter through a 0.2-µm-pore-size filer
- Store ≤1 year at 4°C
FACS buffer
- 500 ml PBS (Corning, cat. no. 21040CM)
- 10 ml (2%) FBS (Sigma, cat. no. F4135)
- Store ≤1 month at 4°C
R10 culture medium
- 500 ml RPMI (Sigma, cat. no. R0883)
- 50 ml (10%) FBS (Sigma, cat. no. F4135)
- 5 ml penicillin/streptomycin (Corning, cat. no. MT30002CI)
- 5 ml (2 mM) L-glutamine (Corning, cat. no. MT25005CI)
- 5 ml (10 mM) HEPES (Corning, cat. no. MT25060CI)
- Store ≤1 month at 4°C
COMMENTARY
Critical Parameters
The controls recommended in this protocol are critical for the interpretation of results. Pre-mixing separately labeled peptide-pulsed and unpulsed cells and comparing increasing E:T ratios against an E:T of 0:1 as a control allows the measurement of titratable peptide-specific killing without confounding effects of nonspecific cell death or errors in pipetting or cell counting.
We have optimized the protocol for use with 25,000-150,000 target cells per well, but to limit sample use, we have attempted using as few as 10,000 target cells per well with success. Because peptide stimulation downregulates TCR from the surface of effector cells, the incubation time after stimulation has been optimized. We and others have found that TCR re-expression as well as perforin and granzyme B expression are maximal at day 6 post-stimulation (Migueles et al., 2020), allowing the assessment of maximal secondary cytotoxicity. We recommend the inclusion of pHLA multimer staining in a control well to distinguish between biological (e.g., lack of responsiveness to stimulation) and technical (e.g., responsiveness but suboptimal TCR re-expression) reasons for a negative result. Including this control also allows the option of normalizing secondary cytotoxicity for the size of the pre- or post-expansion antigen-specific CD8+ T cell pool to calculate the antigen-specific E:T ratio and measure per-cell in addition to endpoint cytotoxicity, which is influenced by both the size and the functional capacity of the response.
EASEA was optimized to measure responses against individual HLA-optimal peptides of 8-12 amino acids in length; however, it may also be used with pooled antigenic peptides. For longer peptides that require endogenous processing and presentation, longer incubation times with target cells may be required before coculture. In our experience, a concentration of 100 nM peptide yields superior responsiveness and TCR re-upregulation by day 6 relative to higher or lower concentrations. At this concentration, a peptide washout step was not necessary to achieve maximal TCR re-expression. Our protocol calls for overloading target cells with 10 µM of peptide during pulsing, but this concentration may be reduced and/or replaced by strategies to endogenously express cognate antigen via infection or transfection. In addition, the pulsing concentration may be titrated experimentally to measure the antigen sensitivity of secondary cytotoxic CD8+ T cell responses, which recent evidence suggests may be particularly important for the evaluation of vaccine-induced responses (Migueles et al., 2023). Although we have optimized the assay using autologous target cells, it is also possible to use target cell lines provided that they express the relevant HLA(s) for killing by the CD8+ T cell response(s) of interest; this may be useful if it is desirable to control for inter-subject heterogeneity in target cell properties that may influence susceptibility to cytolysis. If the primary target cells used may contain replication-competent virus expressing the peptide(s) of interest, antivirals may be included in the target cell culture to prevent viral spread among target cells during expansion, which may otherwise reduce assay sensitivity by enabling antigen presentation in unpulsed target cells.
As with most functional assays involving in vitro cell culture, we have observed that lot-specific variations in the serum added to culture medium can impact results. Therefore, we suggest only comparing results obtained using the same serum lot and/or pre-screening serum lots for comparable performance when switching to a new lot. In our experience, it is sufficient to pre-screen new serum lots using the CFSE dilution and viability readouts on whole peripheral blood mononuclear cells. The suggested flow cytometry panel can be adjusted to replace fluorophores based on available equipment and/or cost limitations. The assay may also be modified to assess immediate ex vivo cytotoxicity, such as for NK cells (Clayton et al., 2021), and elimination of virus-infected target cells and/or non-CD4+ T cell targets, as demonstrated for CD8+ T cell elimination of HIV-infected macrophages (Clayton et al., 2018).
Troubleshooting
Advice for troubleshooting problems that could potentially arise during the assay is summarized in Table 1.
| Problem | Possible cause | Solution |
|---|---|---|
| No or poor killing | Response is poorly functional | Can be confirmed using CFSE dilution, pHLA multimer surface staining, and perforin and granzyme B intracellular staining on day 6 |
| Suboptimal TCR re-expression after proliferation | Wash out peptide from effector cells after day 3 to prevent re-stimulation, modify peptide concentration, and/or modify length of incubation | |
| Insufficient antigen-specific effectors | Increase effector cell number per well and/or decrease target cell number per well |
Understanding Results
Memory CD8+ T cell responses with robust secondary cytotoxic potential will show greater specific elimination of autologous target cells displaying their cognate peptide(s). Conversely, T cell responses with poor cytotoxic potential yield little to no specific elimination. For example, in the context of HIV infection, spontaneous control of viremia is strongly associated with the proliferative and cytolytic potential of antigen-specific CD8+ T cell responses against nonmutated epitopes (reviewed in Collins et al., 2020). EASEA provides data on proliferation via CFSE dilution and the subsequent specific elimination of autologous peptide-pulsed target cells. We recently used EASEA to reveal a longitudinal decline in the cytotoxic potential of antigen-specific CD8+ T cells preceding aborted spontaneous control of HIV viremia using samples from peripheral blood and lymph node excisional biopsies (Collins et al., 2021).
Here we provide example data for antigen-specific responses with different cytotoxic potential. A suggested analytical gating scheme is provided (Fig. 2A). The extent of proliferation is measured by CFSE dilution relative to an unstimulated negative control, and surface TCR-pHLA binding can optionally be assessed using pHLA multimer staining (Fig. 2B). Specific elimination of peptide-loaded target cells is measured by residual frequency of dye-labeled target cells compared to a negative control in which effector cells were not co-cultured (Fig. 2C). The percentage of eliminated target cells (Fig. 2D) at each E:T ratio can be calculated as 100*(1–[residual pulsed target frequency at E:T 1, 2, 4, or 8/pulsed target frequency at E:T 0]). Elimination across a range of E:T ratios can be quantified by area-under-curve (AUC) analysis (Fig. 2E) to enable summary comparisons between responses.
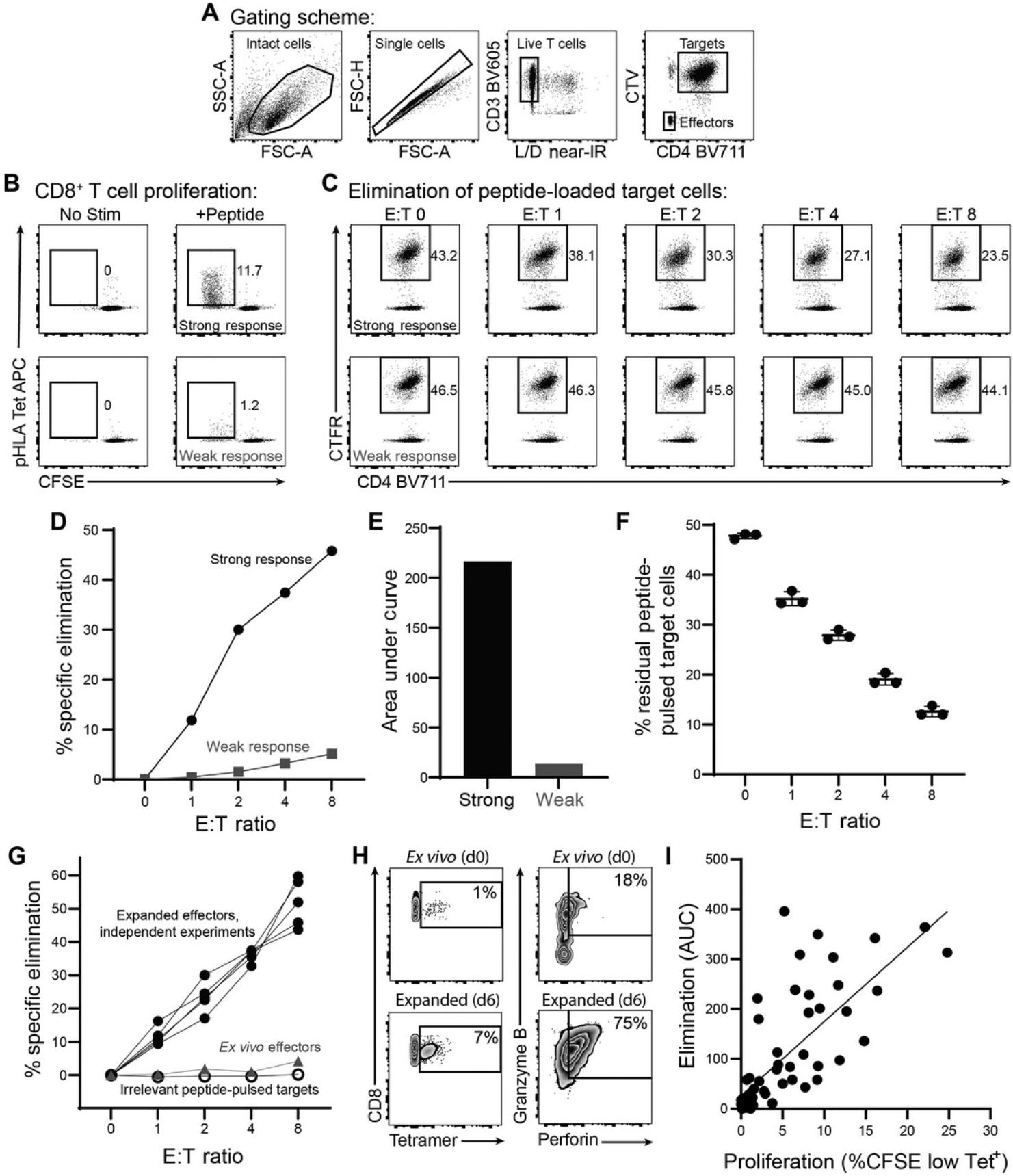
Results are highly reproducible across replicate wells (Fig. 2F) and independent replicate experiments (Fig. 2G). Specificity is internally controlled by measuring the selective elimination of peptide-pulsed relative to unpulsed cells. In these examples, target cells pulsed with an irrelevant peptide, as an additional control, were not eliminated by cognate peptide-expanded cells, further demonstrating specificity (Fig. 2G). Cognate peptide-pulsed cells were poorly eliminated by unexpanded ex vivo effector cells from the same donor, demonstrating the importance of peptide re-stimulation for assessment of secondary cytolytic potential (Fig. 2G). This was attributable to increases in both the number and expression of cytolytic effectors perforin and granzyme B among antigen-specific effector cells (Fig. 2H).
EASEA is designed to capture the combined ability of antigen-specific CD8+ T cells to proliferate and eliminate target cells, functions that are mechanistically related (Migueles et al., 2002). Consistent with this notion, we observed a strong correlation between CD8+ T cell proliferation and specific elimination of autologous peptide-loaded target cells (Fig. 2I). Notably, this correlation is not absolute, and there can be responses with relatively strong elimination despite fewer expanded antigen-specific effectors, as well as weaker elimination despite more numerous expanded antigen-specific effectors, suggesting that additional factors beyond the number and expansion capacity of antigen-specific effector cells influence relative cytotoxicity, consistent with recent literature (Rudloff et al., 2023). Because E:T ratios are calculated using total T cells, they—importantly—do not represent the antigen-specific E:T ratios. Although not required for endpoint measurement of secondary cytotoxic potential, elimination results may be further normalized for relative quantities of antigen-specific effector cells before or after expansion in order to assess relative differences in per-cell elimination capacity.
Time Considerations
The assay spans 6 days and includes ∼10 hr of hands-on time, as indicated in the protocol.
Acknowledgments
We are grateful to Bridget Coffey, Micayla George, James Chen, Nishant Singh, and Alicja Piechocka-Trocha for technical assistance and advice; to the Ragon Institute clinical team for participant recruitment; to Mary Carrington for HLA genotyping; to Michael Waring and the Ragon Institute flow cytometry core facility for instrumentation; and to Ashok Khatri and the Massachusetts General Hospital peptide core facility for reagents. This work was supported by funding from the Howard Hughes Medical Institute and by U.S. National Institutes of Health R01 AI149704 and L30 AI126472. Funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Author Contributions
David R. Collins : Conceptualization; formal analysis; funding acquisition; investigation; methodology; project administration; supervision; visualization; writing—original draft; writing—review and editing. Mpho J. Olatotse : Investigation; writing—review and editing. Zachary J. Racenet : Investigation; writing—review and editing. Umar Arshad : Investigation; writing—review and editing. Elif Çakan : Investigation; validation; writing—review and editing. Gaurav D. Gaiha : Conceptualization; methodology; writing—review and editing. Kiera L. Clayton : Conceptualization; methodology; writing—review and editing. Bruce D. Walker : Funding acquisition; resources; supervision; writing—review and editing.
Conflict of Interest
All authors declare that no conflicts of interest exist.
Open Research
Data Availability Statement
The data that support this protocol are available in this article.
Literature Cited
- Brunner, K. T., Mauel, J., Cerottini, J. C., & Chapuis, B. (1968). Quantitative assay of the lytic action of immune lymphoid cells on 51-Cr-labelled allogeneic target cells in vitro; inhibition by isoantibody and by drugs. Immunology , 14(2), 181–196. https://www.ncbi.nlm.nih.gov/pubmed/4966657
- Clayton, K. L., Collins, D. R., Lengieza, J., Ghebremichael, M., Dotiwala, F., Lieberman, J., & Walker, B. D. (2018). Resistance of HIV-infected macrophages to CD8+ T lymphocyte–mediated killing drives activation of the immune system. Nature Immunology , 19(5), 475–486. https://doi.org/10.1038/s41590-018-0085-3
- Clayton, K. L., Mylvaganam, G., Villasmil-Ocando, A., Stuart, H., Maus, M. V., Rashidian, M., Ploegh, H. L., & Walker, B. D. (2021). HIV-infected macrophages resist efficient NK cell-mediated killing while preserving inflammatory cytokine responses. Cell Host & Microbe, 29(3), 435–447.e439. https://doi.org/10.1016/j.chom.2021.01.006
- Collins, D. R., Gaiha, G. D., & Walker, B. D. (2020). CD8+ T cells in HIV control, cure and prevention. Nature Reviews Immunology , 20(8), 471–482. https://doi.org/10.1038/s41577-020-0274-9
- Collins, D. R., Hitschfel, J., Urbach, J. M., Mylvaganam, G. H., Ly, N. L., Arshad, U., Racenet, Z. J., Yanez, A. G., Diefenbach, T. J., & Walker, B. D. (2023). Cytolytic CD8+ T cells infiltrate germinal centers to limit ongoing HIV replication in spontaneous controller lymph nodes. Science Immunology , 8(83), eade5872. https://doi.org/10.1126/sciimmunol.ade5872
- Collins, D. R., Urbach, J. M., Racenet, Z. J., Arshad, U., Power, K. A., Newman, R. M., Mylvaganam, G. H., Ly, N. L., Lian, X., Rull, A., Rassadkina, Y., Yanez, A. G., Peluso, M. J., Deeks, S. G., Vidal, F., Lichterfeld, M., Yu, X. G., Gaiha, G. D., Allen, T. M., & Walker, B. D. (2021). Functional impairment of HIV-specific CD8+ T cells precedes aborted spontaneous control of viremia. Immunity , 54(10), 2372–384.e2377. https://doi.org/10.1016/j.immuni.2021.08.007
- Fauce, S. R., Yang, O. O., & Effros, R. B. (2007). Autologous CD4/CD8 co-culture assay: A physiologically-relevant composite measure of CD8+ T lymphocyte function in HIV-infected persons. Journal of Immunological Methods , 327(1-2), 75–81. https://doi.org/10.1016/j.jim.2007.07.017
- Fischer, K., Andreesen, R., & Mackensen, A. (2002). An improved flow cytometric assay for the determination of cytotoxic T lymphocyte activity. Journal of Immunological Methods , 259(1-2), 159–169. https://doi.org/10.1016/s0022-1759(01)00507-5
- Jerome, K. R., Sloan, D. D., & Aubert, M. (2003). Measurement of CTL-induced cytotoxicity: The caspase 3 assay. Apoptosis , 8(6), 563–571. https://doi.org/10.1023/A:1026123223387
- Liu, L., Chahroudi, A., Silvestri, G., Wernett, M. E., Kaiser, W. J., Safrit, J. T., Komoriya, A., Altman, J. D., Packard, B. Z., & Feinberg, M. B. (2002). Visualization and quantification of T cell–mediated cytotoxicity using cell-permeable fluorogenic caspase substrates. Nature Medicine , 8(2), 185–189. https://doi.org/10.1038/nm0202-185
- Migueles, S. A., Laborico, A. C., Shupert, W. L., Sabbaghian, M. S., Rabin, R., Hallahan, C. W., van Baarle, D., Kostense, S., Miedema, F., McLaughlin, M., Ehler, L., Metcalf, J., Liu, S., & Connors, M. (2002). HIV-specific CD8+ T cell proliferation is coupled to perforin expression and is maintained in nonprogressors. Nature Immunology , 3(11), 1061–1068. https://doi.org/10.1038/ni845
- Migueles, S. A., Nettere, D. M., Gavil, N. V., Wang, L. T., Toulmin, S. A., Kelly, E. P., Ward, A. J., Lin, S., Thompson, S. A., Peterson, B. A., Abdeen, C. S., Sclafani, C. R., Pryal, P. F., Leach, B. G., Ludwig, A. K., Rogan, D. C., Przygonska, P. A., Cattani, A., Imamichi, H., … Connors, M. (2023). HIV vaccines induce CD8+ T cells with low antigen receptor sensitivity. Science , 382(6676), 1270–1276. https://doi.org/10.1126/science.adg0514
- Migueles, S. A., Rogan, D. C., Gavil, N. V., Kelly, E. P., Toulmin, S. A., Wang, L. T., Lack, J., Ward, A. J., Pryal, P. F., Ludwig, A. K., Medina, R. G., Apple, B. J., Toumanios, C. N., Poole, A. L., Rehm, C. A., Jones, S. E., Liang, C. J., & Connors, M. (2020). Antigenic restimulation of virus-specific memory CD8+ T cells requires days of lytic protein accumulation for maximal cytotoxic capacity. Journal of Virology , 94(23), e01595–e01620. https://doi.org/10.1128/JVI.01595-20
- Reuter, M. A., Del Rio Estrada, P. M., Buggert, M., Petrovas, C., Ferrando-Martinez, S., Nguyen, S., Sada Japp, A., Ablanedo-Terrazas, Y., Rivero-Arrieta, A., Kuri-Cervantes, L., Gunzelman, H. M., Gostick, E., Price, D. A., Koup, R. A., Naji, A., Canaday, D. H., Reyes-Terán, G., & Betts, M. R. (2017). HIV-specific CD8+ T cells exhibit reduced and differentially regulated cytolytic activity in lymphoid tissue. Cell Reports , 21(12), 3458–3470. https://doi.org/10.1016/j.celrep.2017.11.075
- Rudloff, M. W., Zumbo, P., Favret, N. R., Roetman, J. J., Detrés Román, C. R., Erwin, M. M., Murray, K. A., Jonnakuti, S. T., Dündar, F., Betel, D., & Philip, M. (2023). Hallmarks of CD8+ T cell dysfunction are established within hours of tumor antigen encounter before cell division. Nature Immunology , 24(9), 1527–1539. https://doi.org/10.1038/s41590-023-01578-y
- Russell, J. H., & Ley, T. J. (2002). Lymphocyte-mediated cytotoxicity. Annual Review of Immunology , 20, 323–370. https://doi.org/10.1146/annurev.immunol.20.100201.131730
- Saez-Cirion, A., Shin, S. Y., Versmisse, P., Barre-Sinoussi, F., & Pancino, G. (2010). Ex vivo T cell-based HIV suppression assay to evaluate HIV-specific CD8+ T-cell responses. Nature Protocols , 5(6), 1033–1041. https://doi.org/10.1038/nprot.2010.73
- Wherry, E. J. (2011). T cell exhaustion. Nature Immunology , 12(6), 492–499. https://doi.org/10.1038/ni.2035
- Yang, O. O., Kalams, S. A., Trocha, A., Cao, H., Luster, A., Johnson, R. P., & Walker, B. D. (1997). Suppression of human immunodeficiency virus type 1 replication by CD8+ cells: Evidence for HLA class I-restricted triggering of cytolytic and noncytolytic mechanisms. Journal of Virology , 71(4), 3120–3128. https://doi.org/10.1128/JVI.71.4.3120-3128.1997

