Environmental DNA (eDNA) COI PCR Amplification and Gel Electrophoresis Protocol
Alexandria B Boehm, Meghan M. Shea
Abstract
This is a protocol for amplifying DNA extracts via PCR using broad COI primers (Leray et al., 2013) and confirming successful amplification via gel electrophoresis.
This protocol originates with environmental DNA samples collected onto 0.22 µm capped Sterivex filters, e.g. through this sampling and filtration protocol:
Coastal Environmental DNA Sampling & Gravity Filtration ProtocolSamples are then extracted, e.g. through this extracted protocol, before proceeding through the current amplification protocol:
DNA Extraction Protocol from Sterivex Filters Acknowledgements & Attributions:
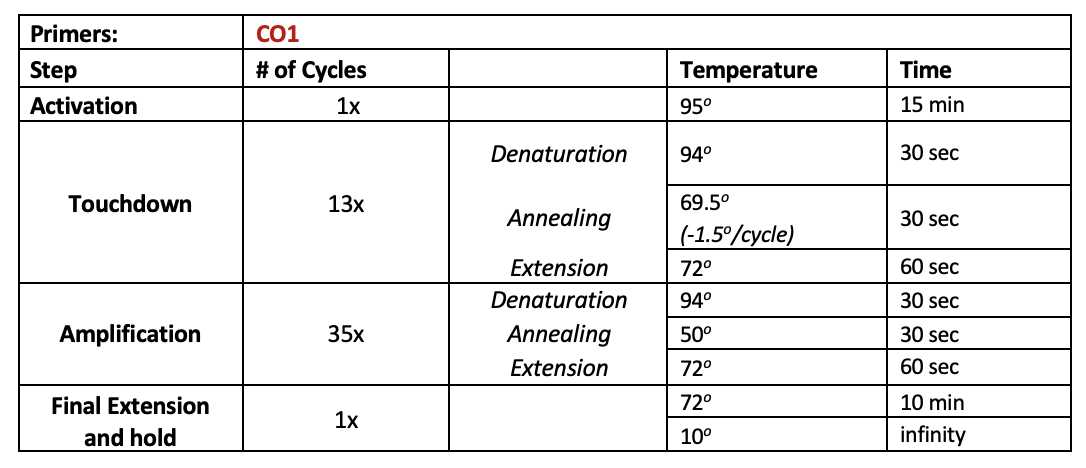
The PCR protocol is adapted from Curd et al. (2019; see Appendix 6) and the detailed first PCR protocol developed by CALeDNA (by Teia Schweizer, Emily Curd, and Rachel Meyer).
The gel electrophoresis protocol is adapted from an Agarose Gel Electrophoresis & Gel Visualization Protocol from the Boehm Lab (by Blythe Layton and edited by Wiley Jennings, Eily Andruszkiewicz Allan, and Winnie Zambrana), a 16S rRNA gene V4 Amplicon Sequencing Protocol from the Nelson Lab (updated by Lauren Kennedy based on the Schloss Lab V4 Protocol) and the detailed Gel Electrophoresis protocol developed by CALeDNA (by Teia Schweizer, Emily Curd, and Rachel Meyer).
We are grateful to the authors and editors of all the protocols above for creating and maintaining such helpful resources.
Attachments
Steps
Set-Up
Wipe down 10 µL pipette with 70% ethanol and RNase Away. UV for 10 minutes on each side
Wipe down a staging area near the PCR hood with 10% bleach, 70% ethanol, and RNase Away
Wipe down ice bin with 10% bleach, 70% ethanol, and RNase away, fill with ice, and remove DNA samples, PCR-grade water, Qiagen Multiplex Mix, and primers from the freezer to thaw. Place bin in staging area near PCR hood
Clean inside of PCR hood (including tube racks, etc.) with 10% bleach, 70% ethanol, and RNase Away
Wipe down PCR hood pipettes with 70% ethanol and RNase Away
Clean any items that need to go into the PCR hood and can be UVed with 10% bleach, 70% ethanol, and RNase Away and then place into PCR hood, including:
- Lab marker
- Bag of 8-well tube strips
- Bag of microcentrifuge tubes
- Strip tube holder
- Any new tip boxes (make sure you have enough in the hood for the rest of the protocol)
With PCR hood cover in place, run UV light for 10 minutes
While PCR hood is UVing, wipe down a vortex, centrifuge, work bench, and box of 10 µL pipette tips with 10% bleach, 70% ethanol, and RNase Away
Once UV timer on PCR hood is done, remove the cover and turn on the regular light
Once reagents have thawed, gently vortex and wipe down the tubes with RNase Away before bringing into the hood
Place a Kimwipe dampened with RNase Away just outside the hood to clean tubes you bring in and out of the hood
Set up your diagram of how you're organizing your samples and calculate the total amount of PCR reagents needed for all samples, e.g. using the attached Sample PCR Calculation Spreadsheet
PCR Preparation
Change gloves before beginning to work in the hood
Label tube strips with relevant information, including:
- Tube number
- Initials
- Date
Add primers and PCR-grade water to microcentrifuge tube(s) according to your combined reagent recipe (see attached Sample PCR Calculation Spreadsheet), remove from hood, vortex vigorously, wipe with RNase Away, and return to hood
Add Qiagen Multiplex Mix to microcentrifuge tube(s) according to your master mix recipe, remove from hood, vortex VERY gently, wipe with RNase Away, and return to hood
Using a pipette, aliquot 24 µL of master mix to each strip tube
Adding Template DNA
Remove the aliquoted master mix and put on bench
Vortex each DNA sample and pipette 1 µL into the appropriate tube strip. Mix the DNA and master mix solution by pipetting up and down
Thermocycling
Place your samples in to the thermocycler, and double check that all tubes are fully sealed
The thermocycler will take between 2 and 4 hours to complete the program. While the thermocycler is running, prepare for gel electrophoresis (below)
When the thermocycler has finished running, remove your tubes and turn off thermocycler
Spin your PCR tubes down using a centrifuge or microcentrifuge specialized for plates or tubes and label. Make sure the labels are clear.
Gel Electrophoresis Preparation
Weigh out 1.9 g of agarose and measure out 125 ml of 1X TAE for a 1.5% gel
Heat agarose in TAE in microwave until the agar is completely dissolved. You will know the agarose is completely dissolved when there are no specks floating in the TAE.
Allow the solution to cool to just above room temperature. Placing the flask on a few folded paper towels will prevent the agarose on the bottom from solidifying too soon.
Once cool, add 12.5 ul of GelRed per 125 mL of gel (want it to be 1X final concentration). Gently swirl until GelRed is dissipated evenly.
Fit the casting tray in the box
Pour the agarose into the casting tray and place the combs in the tray. Use the “fatter” side of the combs to make it easier to pipet into.
Allow the gel to solidify
Remove the tray and put in running position with the combs closer to the negative (black) electrode
Fill the box with TAE buffer until it just covers the gel
Carefully remove the combs
Decide the subset of samples to visualize via gel electrophoresis to confirm successful amplification
Gel Electrophoresis
Load 5 µl of DNA ladder (at concentrations specified by the manufacturer) into the wells on either end of the row of samples
Dot 2 µl of loading dye (one dot per PCR product) onto a piece of parafilm
Mix 4 µl of PCR product with the dye by pipetting up and down
Load the entire ~6 µl sample into a well, ejecting the sample slowly and carefully to avoid spillage or bubbles
Run gel at 110 volts for 60 minutes
Visualize and photograph gel using UV light box, GelDoc, etc
If visualizing with GelDoc, use the following steps:
Use a paper towel to open the drawer of the GelDoc.
Remove the tray from the gel box and carefully slide the gel onto the glass surface of the drawer
Remove one glove, close the drawer and open the main door of the GelDoc. Open the QuantityOne software and select File -> GelDoc XR.
Turn on the white lamp by pressing the Epi White button and click “Live/Focus”
Use your gloved hand to center the gel
Close the door and turn on the UV lamp (Trans UV button)
Take a picture by clicking either “Auto Expose” or “Manual Acquire”, then “Freeze”. Adjust the exposure length as necessary until you are satisfied with the image.
Click “Save,” then “Print.”
Dispose of the gel properly and clean the glass surface of the GelDoc with a paper towel.