Density Gradient Centrifugation-Independent Purification of Human Basophils
Natalie Gray, Natalie Gray, Daniela Wiebe, Daniela Wiebe, Tobias Weihrauch, Tobias Weihrauch, Ulrike Raap, Ulrike Raap, Maren M. Limberg, Maren M. Limberg
basophil activation test
basophil purification
calcium flux analysis
flow cytometry
negative immunomagnetic selection
viability
Abstract
Basophils represent the rarest type of granulocyte in human peripheral blood. Thus, researching basophils has historically been challenging and has often been reliant on enrichment protocols using density gradient centrifugation. This article describes a novel, fast, and cost-effective method to purify highly viable human basophils from peripheral blood through negative immunomagnetic selection, foregoing the density centrifugation step in the Basic Protocol. The technique is easy to use and consistently produces purities >96%. Furthermore, the Support Protocols describe procedures to determine basophil yield, purity, and viability, and how to investigate functional activity of the purified basophils through flow cytometry and visualize the basophils through microscopy. © 2024 The Authors. Current Protocols published by Wiley Periodicals LLC.
Basic Protocol : Gradient centrifugation-independent basophil isolation
Support Protocol 1 : Flow cytometry staining to assess basophil yield, purity, and viability
Support Protocol 2 : Giemsa staining
Support Protocol 3 : Calcium flux analysis
Support Protocol 4 : Basophil activation test
INTRODUCTION
Basophil granulocytes are the rarest type of leukocyte in human peripheral blood. They make up <1% of all white blood cells with ∼2–8 × 104 cells being present per ml of blood (Schroeder & Bieneman, 2016). Their scarcity has presented challenges in researching basophils, but enrichment protocols using density gradient centrifugation procedures have provided a method to isolate sufficient numbers of these cells for research. However, these methods have their drawbacks, as the medium used for the density gradient centrifugation can negatively affect the yield and viability of isolated basophils. Additionally, to achieve purities >96%, further purification steps using magnetic bead-based selection kits are required, increasing both the cost and time requirements of the protocols.
We present a novel time-efficient method using negative immunomagnetic selection to directly isolate highly pure human basophils from peripheral blood, foregoing the density gradient centrifugation step (Basic Protocol). The manufacturers’ protocol was modified to use a lower amount of antibody cocktail and magnetic beads, therefore reducing the cost, but still ensuring cell purities to be consistently >96%. We have used this method for publishing our research on basophils in Limberg et al. (2023) and Gray et al. (2022). Furthermore, we describe a flow cytometry staining protocol to assess basophil yield, purity, and viability (Support Protocol 1), as well as a staining protocol to visually inspect the purified basophils (Support Protocol 2). For evaluation of the basophils’ functional integrity, we also provide a protocol to observe changes in intracellular Ca2+ levels (Support Protocol 3) and describe how to perform the basophil activation test (BAT) to investigate degranulation status (Support Protocol 4).
CAUTION : Biosafety practices should be followed when working with human blood, purified cells, or other potentially infectious agents. These include using filter tips for pipettes that are single use, and wearing personal protective equipment, such as gloves and eye protection. If desired handling can be performed in a laminar flow bench, though this is not strictly necessary to protect the cells, as basophils do not replicate and therefore cannot be cultured for long periods of time. After completion of the experiments, all reagents and surfaces must be decontaminated appropriately.
NOTE : When working with human samples, approval of the responsible ethics committee and appropriate informed consent must be obtained before the commencement of sample collection and experiments.
Basic Protocol: GRADIENT CENTRIFUGATION-INDEPENDENT BASOPHIL ISOLATION
This protocol describes a quick and optimized method to isolate human basophils from whole blood through negative-immunomagnetic selection.
Materials
-
Peripheral blood, freshly drawn, no older than 4 hr, collected in 9-ml EDTA K3E preloaded monovettes (Sarstedt, cat. no. 02.1066.001)
- If anticoagulants other than EDTA are used when collecting blood samples, EDTA at a final concentration of 3 mM must be added to the whole blood sample prior to isolation to ensure proper performance of the kit.
-
EasySep direct human basophil isolation kit (STEMCELL Technologies, cat. no. 19667)
-
Phosphate-buffered saline (PBS)-EDTA buffer (see recipe) or EasySep buffer (STEMCELL Technologies, cat. no. 20144)
-
13-ml, 95-mm × 16.8-mm, polystyrene, round-base tubes (Sarstedt, cat. no. 55.468.005)
-
3-ml single use Pasteur pipettes (Ratiolab, cat. no. 2600111)
-
“The Big Easy” EasySep magnets (STEMCELL Technologies, cat. no. 18001)
-
50-ml polypropylene conical tubes (Sarstedt, cat. no. 62.547.254)
-
Tabletop centrifuge (Eppendorf centrifuge 5810R)
NOTE : The amount of blood that can be processed depends on the number of available “The Big Easy” Magnets. One magnet can isolate basophils from 2 to 6 ml of whole blood. Other magnet sizes are available from STEMCELL, but the protocol has not been tested with these.
Basophil purification
1.Collect blood by venipuncture into the EDTA preloaded monovettes. To reduce hemolytic forces on cells, minimal force should be used when retracting the plunger.
2.Gently invert the monovettes 5 times to mix the EDTA with the blood.
3.Transfer 6 ml blood into each round-base tube.
4.Vortex the EasySep RapidSpheres for ≥30 s. Visually check if settled beads at the bottom of the tube have been resuspended. If this is not the case, continue vortexing until no sedimentation remains visible.
5.Add 150 µl EasySep antibody cocktail and 150 µl RapidSpheres to each tube and mix by pipetting.
6.Incubate at room temperature for 5 min.
7.Add 5700 µl PBS-EDTA buffer to a total volume of 12 ml and mix by pipetting.
8.Place round base tube into the “The Big Easy” magnet and incubate at room temperature for 5 min.
9.In one continuous motion and without removing the tube from the magnet, pour the contents of the tube into a new round base tube until the stream of liquid naturally interrupts and begins to drip.
10.Add 75 µl RapidSpheres to the tube and mix evenly.
11.Incubate at room temperature for 5 min.
12.Place tube into the “The Big Easy” magnet and incubate at room temperature for 5 min.
13.Without removing the tube from the magnet, pour the contents into a new tube in one continuous motion.
14.Add 75 µl RapidSpheres to the tube and mix evenly.
15.Incubate at room temperature for 5 min.
16.Place tube into the “The Big Easy” magnet and incubate at room temperature for 5 min.
17.Without removing the tube from the magnet, pour the contents into a new tube in one continuous motion.
18.Add 75 µl Rapid Spheres and 20 µl antibody cocktail to the tube and mix gently.
19.Incubate at room temperature for 5 min.
20.Place tube into the “The Big Easy” magnet and incubate at room temperature for 5 min.
21.Transfer the contents to a 50-ml conical tube.
22.Centrifuge the cell suspension 7 min at 350 × g , room temperature.
23.Remove and discard the supernatant with a serological or Pasteur pipette.
The basophils are now ready to be gently resuspended for further experiments and analysis. We found resuspension densities of 500 to 2500 basophils per µl of medium to work well. The type of medium used for resuspension and the volumes need to be selected according to the type of experiment for which the cells are intended to be used.
Support Protocol 1: FLOW CYTOMETRY STAINING TO ASSESS BASOPHIL YIELD, PURITY, AND VIABILITY
This protocol describes the flow cytometry procedure to determine the yield, purity, and viability of human basophils in the isolated cell fraction. For this, a small volume of cell suspension is removed from the resuspended isolated cells. Yield and purity are assessed using basophil specific FACS-antibodies. Viability is determined through the exclusion of 7-AAD-positive cells.
Materials
-
Purified human basophil suspension (see Basic Protocol)
-
PBS w/o Ca2+ and Mg2+ (ROTH, cat. no. 9143.2)
-
Pacific Blue (PB)-conjugated anti-human CD123 antibody (Biolegend, cat. no. 306044, RRID: AB_2750165)
-
Allophcocyanin (APC)-conjugated anti-human FcεRIα antibody (Biolegend, cat. no. 334612, RRID: AB_10578086)
-
7-AAD staining solution (Miltenyi, cat. no. 130-11-568)
-
1.5-ml microcentrifuge tubes (Sarstedt, cat. no. 72.690.001)
-
Flow cytometer (Beckman Coulter, Cytoflex S)
Flow cytometry staining
1.Prepare two 1.5-ml microcentrifuge tubes.
2.Add 5 µl of the purified, resuspended cell suspension and 45 µl PBS buffer to each tube.
3.Add 0.65 µl PB-conjugated anti-CD123 and 0.65 µl APC-conjugated anti-FcεRIα to the first tube and mix by pipetting.
4.Add 1 µl of 7-AAD staining solution to the second tube and mix by pipetting gently.
5.Incubate both tubes in the dark at room temperature for 10 min.
6.Dilute the stained cell suspensions with 200 µl PBS.
7.Proceed with flow cytometry analysis.
Support Protocol 2: GIEMSA STAINING
This protocol describes the staining procedure used to differentiate different cell types in blood smears. It can also be used to visually confirm the purity of isolated human basophils in cytospins. Nuclei will appear red to violet and granules will be stained dark purple.
Materials
-
Purified human basophils (see Basic Protocol)
-
PBS buffer (see recipe)
-
Methanol (ITW Reagents, cat. no. 221091)
-
Giemsa staining solution (see recipe)
-
Buffer solution, pH 7.2 (see recipe)
-
H2O, ultrapure
-
0.5% acetic acid solution (see recipe)
-
100% ethanol (Merck, cat. no. 1009861000)
-
100% isopropyl alcohol (Merck, cat. no. 33539-IL-M)
-
Neo-Mount mounting medium (Merck, cat. no. 1090160500)
-
Filter cards (Fisher Scientific, cat. no. 5991022)
-
Adhesive microscopy slides (Marienfeld Superior, cat. no. 0810001)
-
Epredia TPX single sample chamber (Fisher scientific, cat. no. A78710018)
-
Shandon Cytoclip stainless steel slide clip (Fisher scientific, cat. no. 12608036)
-
Shandon Cytospin 4 cytocentrifuge (Fisher scientific, cat. no. A78300003)
-
Glass staining trough (Engelbrecht, cat. no. 42460)
-
Hydrophobic barrier pen (Merck, cat. no. Z377821-1EA)
-
Cover slips, 18-mm × 18-mm (VWR, cat. no. 631-1567)
Preparing cytospins
1.Resuspend the basophil pellet with PBS to a density of ∼1000 basophils/µl.
2.Place the filter card between the microscopy slide and the sample chamber so that the hole in the card aligns with the opening of the funnel.
3.Secure the construction with a cytoclip, and place into the cytocentrifuge.
4.Pipette 50 µl basophil suspension into the funnel of the sample chamber.
5.Centrifuge 5 min at 500 × g , room temperature.
6.Remove the sample chambers from the cytocentrifuge and remove the slides from the setup.
7.Air dry slides for 10 min.
Staining cytospins
8.Place cytospins in a methanol-filled glass staining trough and fix slides for 5 min.
9.Allow slides to air dry.
10.Surround the spot where basophils are concentrated on the slides with a hydrophobic barrier pen and briefly air dry.
11.Place cytospins slides on a flat surface.
12.Pipette 200 µl diluted Giemsa staining solution onto the basophil spot.
13.Incubate at room temperature for 30 min.
14.Wash twice in buffer solution for 1 min.
15.Rinse the slides by quickly dipping them 5 times in a trough filled with ultrapure water.
16.Dip the slides in 0.5% acetic acid solution 5 times.
17.Dip the slides in 100% ethanol 5 times.
18.Dip the slides in 100% isopropyl alcohol 5 times.
19.Let slides air dry.
20.Mount slides with 50 µl mounting medium and cover with coverslips.
Support Protocol 3: CALCIUM FLUX ANALYSIS
This protocol describes the procedure to measure changes in intracellular calcium levels within human basophils through flow cytometry. Basophils are preloaded with a calcium-sensitive fluorescent dye, which enables the response to stimuli to be observed in real time by measuring the changes in fluorescence.
Materials
-
Purified human basophils (see Basic Protocol)
-
RPMI 1640 medium, without phenol red (ThermoFisher Scientific, cat. no. 32404014)
-
Fluo-4 AM (ThermoFisher Scientific, cat. no. F14217)
-
Ionomycin (Fisher Scientific, cat. no. 10134232)
-
1.5-ml microcentrifuge tubes (Sarstedt, cat. no. 72.690.001)
-
CO2 incubator (Eppendorf, Galaxy 170S)
-
Flow cytometer (Beckman Coulter, Cytoflex S)
-
Gel loading pipette tips (ThermoFisher Scientific, cat. no. 11367801)
Preparing the cells
1.Resuspend the basophil pellet in 500 µl prewarmed, phenol red-free RPMI medium, for each experimental condition (a total volume of 1000 µl for two conditions; 2000 µl for four conditions, etc.).
2.Prepare one 1.5-ml microcentrifuge tube per experimental condition.
3.Transfer 500 µl basophil suspension into each microcentrifuge tube.
4.Add 1.5 µl Fluo-4 AM to each tube and mix by pipetting thoroughly.
5.Incubate for 20 min at 37°C and 5% CO2.
6.Centrifuge 5 min at 350 × g , room temperature.
7.Remove and discard the supernatant.
8.Resuspend cell pellets in 450 µl prewarmed phenol red-free RPMI medium.
Flow cytometry detection of changes in intracellular calcium levels
9.Place the Fluo-4 loaded basophil suspension into the flow cytometer and start a continuous timeline measurement of the Fluo-4 fluorescence on medium speed (30 µl/min).
10.Measure calcium related fluorescence until a stable baseline is visible on the flow cytometer screen (∼1 min)
11.Add appropriately diluted stimulants in a volume of 50 µl phenol red-free RPMI medium directly into the cell suspension in the microcentrifuge tubes using a gel loading pipette tip.
12.Simultaneously manually boost the flow cytometer uptake speed to the maximum. Hold at the maximum for 2 s and then return the flow rate to medium (30 µl/min).
13.Continue the continuous timeline measurement for several minutes.
Support Protocol 4: BASOPHIL ACTIVATION TEST
This protocol describes the procedure to measure anaphylactic and piecemeal degranulation of basophils in response to different stimuli. CD63 is a marker that is associated with anaphylactic degranulation and is usually not present on resting basophils. CD203c is present on resting basophils in low amounts but is significantly upregulated as a result of piecemeal degranulation. As externalization of CD63 and CD203c on basophils increases with degranulation, flow cytometry analysis of these markers can be used to observe the degranulation status of basophils in response to stimuli.
Materials
-
Purified basophils (see Basic Protocol)
-
RPMI 1640 medium (VWR, cat. no. 392-0426)
-
Stimuli and anti-IgE (Merck, cat. no. I6284) or fMLP (Merck, cat. no. F3506) positive controls
-
BAT master mix (see recipe)
-
PBS buffer w/o Ca2+ and Mg2+ (ROTH, cat. no. 9143.2)
-
1.5-ml microcentrifuge tubes (Sarstedt, cat. no. 72.690.001)
-
Block heater (MyBlock, mini dry bath)
-
Ice
-
Microcentrifuge (Eppendorf, Centrifuge 5425R), 4°C
-
Flow cytometer (Beckman Coulter, Cytoflex S)
Basophil activation test
1.Resuspend the basophil pellet in 90 µl prewarmed, RPMI 1640 for each experimental condition (a total volume of 180 µl for two conditions; 360 µl for four conditions, etc.).
2.Prepare one 1.5-ml microcentrifuge tube per experimental condition.
3.Transfer 90 µl basophil suspension into each microcentrifuge tube.
4.Add 10 µl of appropriately diluted stimuli and mix well by pipetting.
5.Place the microcentrifuge tubes into a cell incubator and incubate 30 min at 37°C and 5% CO2.
Flow cytometry measurement of basophil activation
6.Place the microcentrifuge on ice for 2 min to stop cell activity.
7.Centrifuge 5 min at 350 × g , 4°C.
8.Carefully remove and discard the supernatants.
9.Resuspend the basophil pellets with 50 µl BAT master mix and mix well by pipetting.
10.Incubate for 10 min in the dark at room temperature.
11.Add 200 µl PBS and mix by pipetting.
12.Proceed with flow cytometry analysis.
REAGENTS AND SOLUTIONS
Acetic acid solution, 0.5%
- 95.5 ml ultrapure water
- 0.5 ml glacial acetic acid (Fisher Scientific, cat. no. 11337558)
- Prepare fresh before each staining
BAT master mix
- Per experimental condition:
- 48 µl PBS buffer w/o Ca2+ and Mg2+ (ROTH, cat. no. 9143.2)
- 0.5 µl Pacific Blue anti-human CD123 antibody (Biolegend, cat. no. 306044, RRID: AB_2750165)
- 0.5 µl APC anti-human FcεRIα antibody (Biolegend, cat. no. 334612, RRID: AB_10578086)
- 0.5 µl FITC anti-human CD63 antibody (Biolegend, cat. no. 353006, RRID: AB_10898319)
- 0.5 µl PE anti-human CD203c (Biolegend, cat. no. 324606, RRID: AB_756044)
- Prepare fresh and protect from light
Giemsa staining solution
- 5 ml Giemsa azur eosine methylene blue solution (Merck, cat. no. 1.09204)
- 95 ml staining buffer, pH 7.2 (see recipe)
- Prepare fresh before each staining
PBS buffer
- 1 L ultrapure water
- 9.55 g ROTI PreMix PBS (ROTH, cat. no. 0890.2)
- Mix until the PreMix PBS is completely dissolved
- Store up to 3 months at 4°C
PBS-EDTA buffer
- PBS buffer w/o Ca2+ and Mg2+ (ROTH, cat. no. 9143.2)
- 1 mM EDTA (ROTH, cat. no. 8040.2)
- Store up to 1 month at 4°C
Staining buffer, pH 7.2
- 1 L ultrapure water
- 1 buffer tablet, pH 7.2 to prepare buffer solution according to WEISE (Merck, cat. no. 1.09468)
- Wait until tablet is completely dissolved and mix well
- Store up to 3 months at 4°C
COMMENTARY
Background Information
Basophils are pivotal players in allergic inflammatory diseases, e.g., atopic dermatitis, contact dermatitis, asthma, anaphylaxis and even in autoimmune inflammatory diseases, such as bullous pemphigoid. Morphologically, basophils are 5 to 8 µm in diameter and characteristically possess condensed, segmented nuclei (Stone et al., 2010). Furthermore, basophils contain numerous granules that contain proinflammatory mediators and cytokines, such as leukotriene (LTC4) and histamine (Martelletti et al., 1989). Through the release of immunomodulatory Th2-type cytokines, e.g., IL-4 (MacGlashan et al., 1994), IL-13 (Li et al., 1996), and IL-31 (Raap et al., 2017), they are also able to recruit and modulate the function of other immune cells, such as eosinophils in allergic inflammation (Berger, 2000; Iype & Fux, 2021). Basophils are traditionally activated by IgE-antibodies crosslinking high affinity IgE-receptors (FcεRIα) (Saini et al., 1999), though they can also be activated through other agents, e.g., cytokines and proteases, and possess a multitude of receptors, such as IL-4R, IL-13R, and IL31RA (Siracusa et al., 2013; Wiebe et al., 2023). When performing a basophil activation test in vitro , activation in response to allergens and other stimuli is assessed through flow cytometry analysis of activation markers, such as CD63 and CD203c. This is an important diagnostic tool for diagnosing allergies and basophil activation status (Doña et al., 2021).
Until recently, no human basophil cell culture line existed; research was dependent on freshly isolated cells from donors before each experiment. Even with the recently established cell line (Celprogen, cat. no. 36046-17-T150), purification of basophils from multiple donors is not redundant, as high inter-donor variability cannot be portrayed accurately with solely one cell culture line. Due to their scarcity, relatively large amounts of peripheral blood are required to purify adequate numbers of basophils to perform experiments. Therefore, some labs utilize concentrated buffy coat products from blood donation services to obtain a sufficient number of cells. But, depending on the timing of blood donation, sample processing, and sample collection, freshness of the buffy coats cannot always be guaranteed, potentially causing the short-lived basophils to already enter apoptosis. Moreover, blood donation services do not require donors do disclose all underlying health conditions, such as allergies. This could possibly influence experimental results if basophils from atopic donors are unknowingly used for experiments as basophils from atopic donors have been found to possess higher baseline CD63 externalization levels than those of healthy donors (Liang et al., 2023).
Human basophils are most often isolated from peripheral blood through protocols involving density gradient centrifugation. Different density gradient centrifugation media are available, though Ficoll and Percoll seem to be most commonly used (Gibbs et al., 1997; Shiono et al., 2016). Each medium possesses distinct advantages and disadvantages. Basophil recovery rates using Ficoll appear to be fairly high (Miroli et al., 1986), but as Ficoll poses a potentially toxic environment to cells (Pösel et al., 2012), this method of isolation may not be optimal if the isolated basophils are to be used in assays measuring functional activity. In contrast, though non-toxic, enrichment using Percoll, seems to result in relatively low basophil yields (Kepley et al., 1994; Raghuprasad, 1982). Additionally, enrichment protocols using Ficoll or Percoll necessitate subsequent steps to achieve basophil purities of >96%, which are required if further methods of analysis do not provide the option of labeling cells with identifying markers (e.g., PCR analysis). The additional purification steps not only increase the time until basophils are ready for further assays, but depending on the reagents used, may also significantly increase the cost of the basophil isolation procedure.
By foregoing the density centrifugation step, our direct isolation protocol significantly decreases the time required to purify highly pure human basophils. Additionally, we were also able to reduce the cost per isolation by modifying the amount of isolation cocktail and magnetic selection beads.
Critical Parameters and Troubleshooting
Researchers should be familiar with the safety precautions necessary to handle potentially infectious blood samples. Appropriate personal protective equipment, such as gloves and eye protection, must be used. Additionally, it is recommended to use filter tips while pipetting to avoid cross contamination to other samples. Researchers should also possess basic knowledge of flow cytometry to confidently distinguish between the different cell populations.
The presence of EDTA is vital for the efficacy of the isolation method. If other anti-coagulants are used when collecting blood samples, the appropriate amount (3 mM) of EDTA needs to be added prior to commencement of basophil isolation. Blood samples should be processed within a few hours of collection to ensure that basophil viability is optimal and to ensure the reliability of subsequent experiments.
Basophil numbers can vary greatly depending on the donor. Even yields from the same donor can show variation depending on the day of collection. Though rare, some donors might present with complete basopenia, making the isolation of basophils from blood samples impossible. Unfortunately, even some healthy presenting donors will have virtually no basophils in their peripheral blood. Some conditions, such as type 1 diabetes may also increase the numbers of basophils in donors. This should be taken into consideration when deciding on how much whole blood to isolate for experiments.
As basophils are fairly sensitive cells that are prone to degranulation, it is critical to handle unprocessed blood samples and purified basophils carefully. Blood samples should be transported in a fashion that minimizes vibrations and cell suspension pipetted in a slow and controlled manner, avoiding the formation of air bubbles.
When performing BAT analysis some donors may show high levels of CD63 and CD203c externalization, even in the negative control. This most likely is due to the donors having come in contact with substances they are allergic to. Therefore, if donors have known allergies, BAT analysis should be performed at a time point prior to which no contact with the allergen occurred.
Statistical Analysis
We use the software Prism (GraphPad) to statistically analyze flow cytometry data. Data are tested for normal Gaussian distribution using a Shapiro-Wilk normality test. Depending on how many experimental conditions are investigated, a paired t test or one-way ANOVA may be used.
Understanding Results
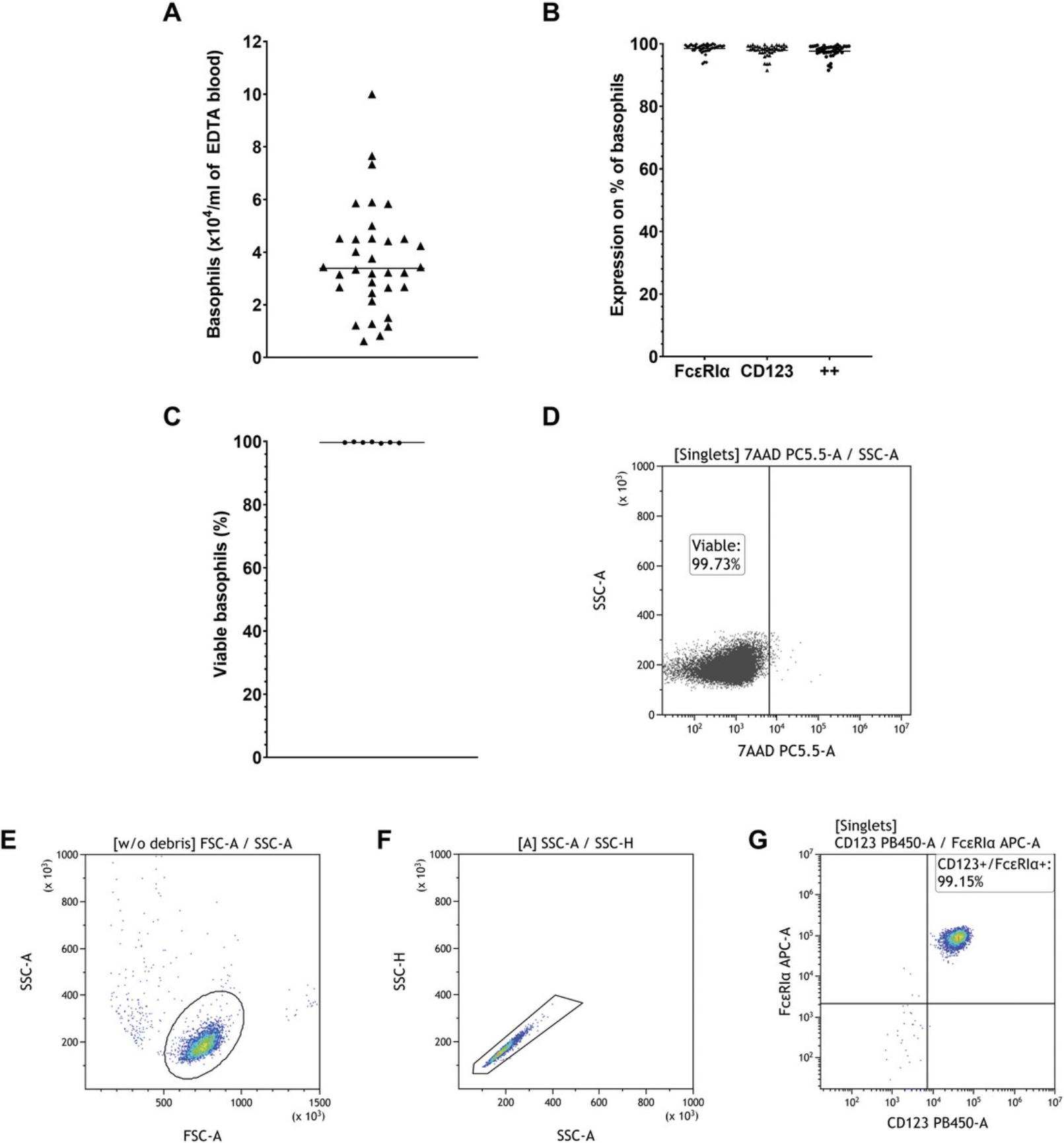
Basophils are the least abundant type of granulocyte, making up only 0.1% to 1% of all leukocytes. Due to inter-donor variability, basophil yield after purification may vary. We typically are able to retrieve 2–6 × 104 (mean = 3.74 × 104) human basophils per ml of whole blood as shown in Figure 1A, though yields above and beyond this range are not rare. Purity of the isolated basophils usually was 96% to 99% (mean = 97.64%) as illustrated in Figure 1B. We identified basophils through flow cytometry by selecting CD123 and FcεRIα double-positive cells. If basophils are purified from the blood samples shortly after donation (no longer than 4 h) we found the viability of the isolated cells, which was measured by the exclusion of 7-AAD positive cells, to be consistently >99% (mean = 99.69%) (Fig. 1C). A representative dot plot of the cell viability measurement is shown in Figure 1D. When analyzing the cells using flow cytometry, it is crucial to only measure single cells, excluding doublets. It can also be beneficial to exclude debris. The gating strategy to identify basophils is illustrated in Figure 1E-G.
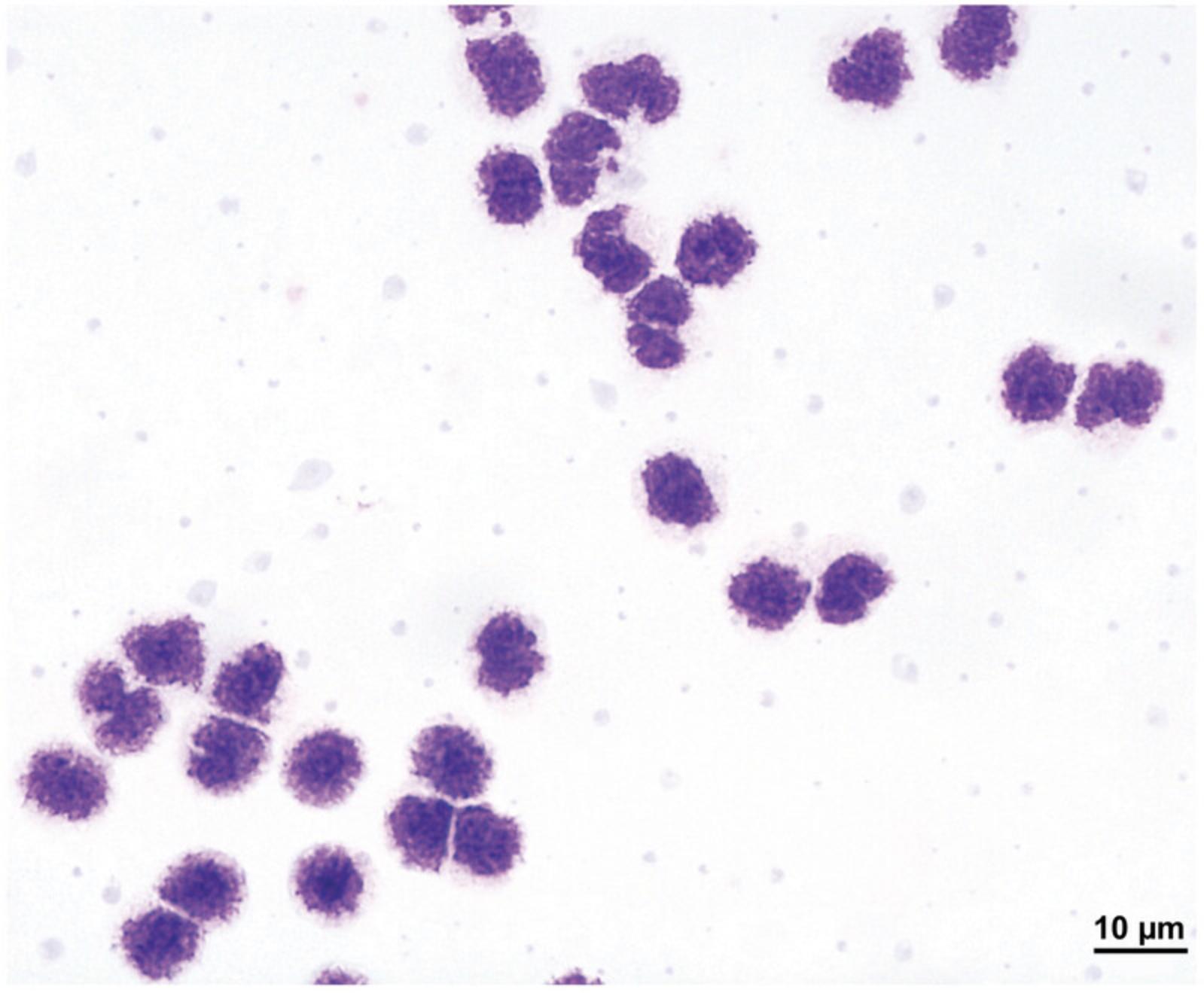
In addition to using flow cytometry analysis, Giemsa staining of cytospins can be used to visually confirm basophil purity in the isolated cell fraction. Basophils are identifiable by their characteristic polymorphonuclear nucleus and numerous granules, which will be stained dark purple. Contaminating cells are easily identifiable by their diverging appearance and coloration. A representative image of highly pure Giemsa-stained basophils at 100× magnification can be seen in Figure 2.
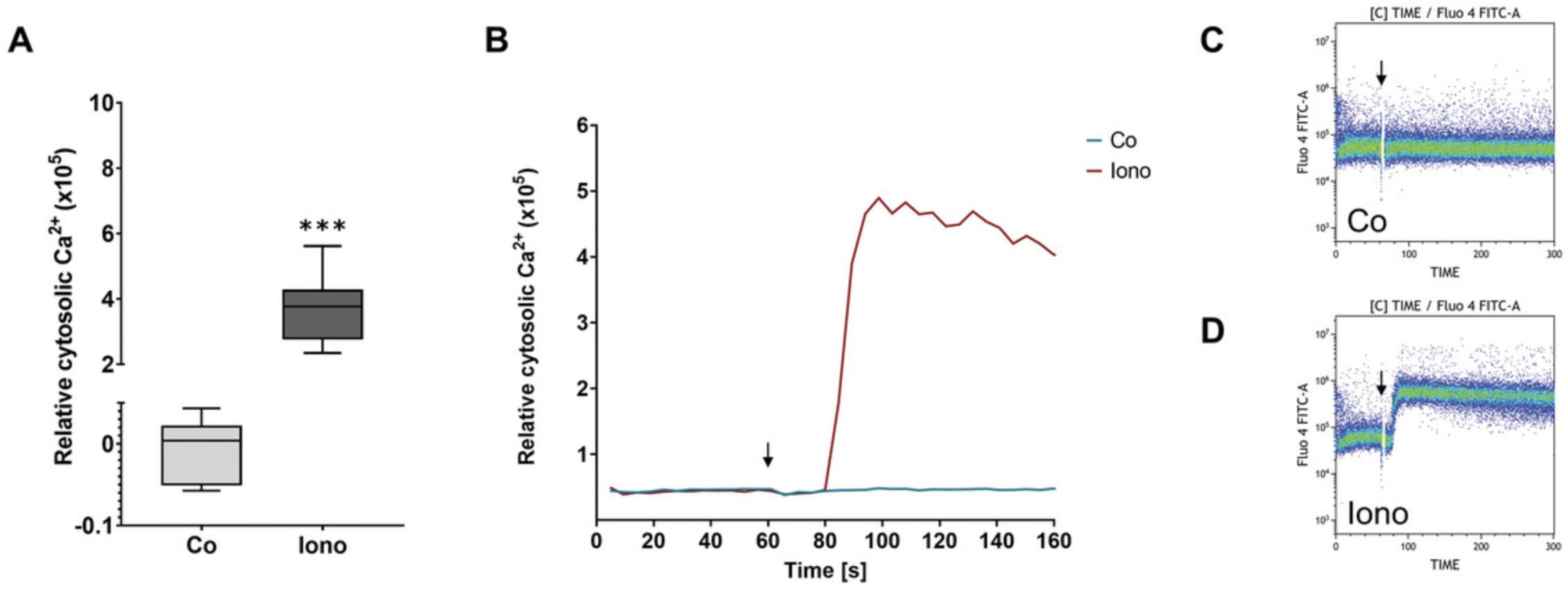
The functional integrity of the isolated human basophils can be tested by observing changes in intracellular calcium levels in response to stimuli. For this, basophils are loaded with the calcium-sensitive fluorescent dye Fluo-4 AM, and stimuli are added while the fluorescence is being measured in the flow cytometer. Typical changes in intracellular Ca2+ levels in response to 500 mM ionomycin are depicted in Figure 3A. To obtain these data, relative cytosolic Ca2+ before stimulation was subtracted from relative cytosolic Ca2+ after stimulation. A representative image of calcium flux over time after application of a negative (Co) and positive control (Iono) during the application of stimuli (at 60 s), is shown in Figure 3B, and the corresponding cytograms depicted in Figure 3C and 3D. As ionomycin is an ionophore, cell response and resulting calcium flux is very high. Cell response to other stimuli, e.g., cytokines and mediators, will be more moderate but still visible.
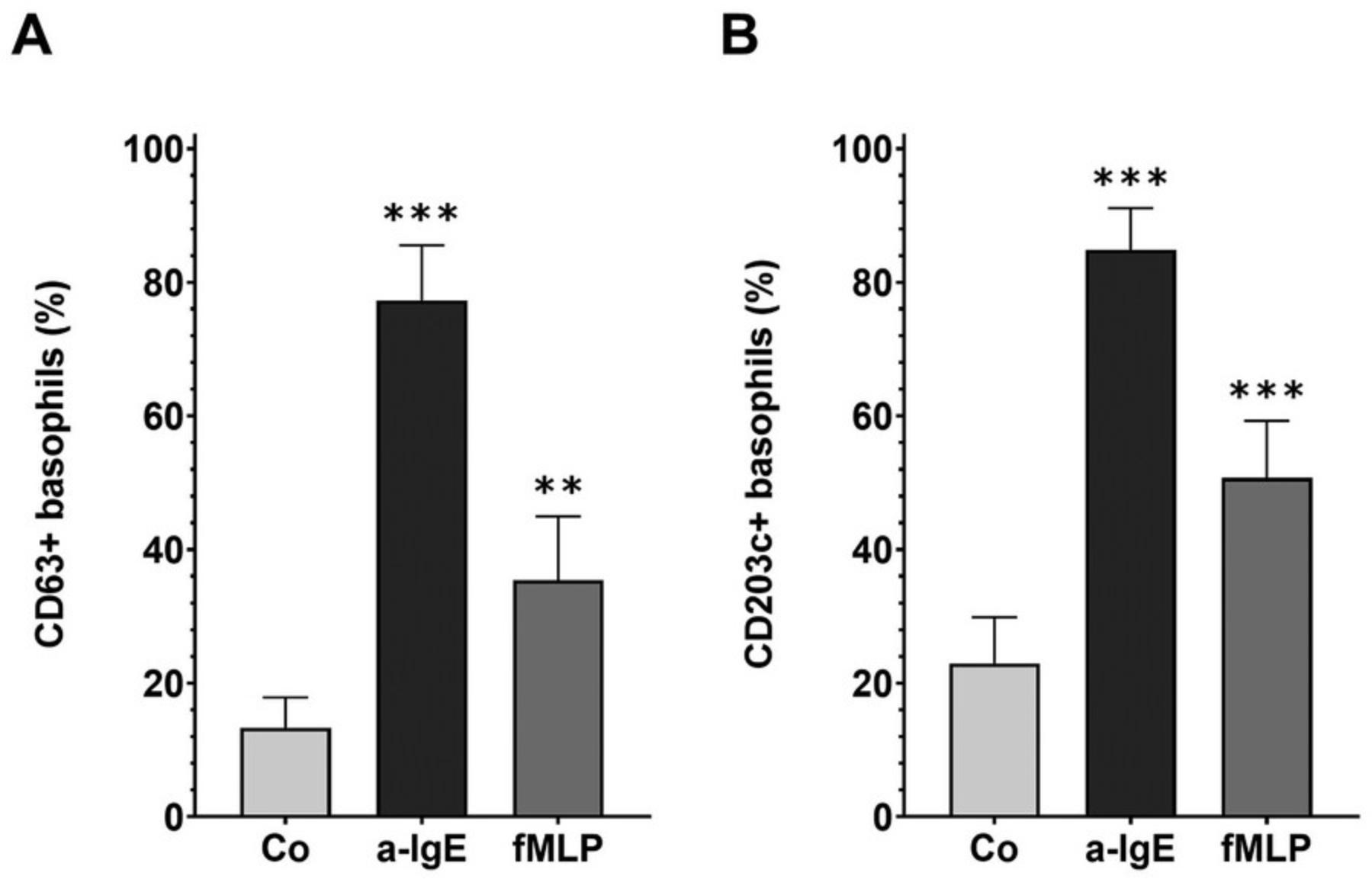
Another test to determine the functional integrity of purified human basophils is the basophil activation test (BAT). This test investigates the externalization of the degranulation markers CD63 and CD203c in response to stimuli, and therefore basophils’ main biological function. Basophils are selected as shown in Figure 1E-G and the activation markers also measured by flow cytometry. Gates to identify positive cells should be set according to fluorescence minus one (FMO) controls, and the specificity of antibodies tested with the appropriate isotype controls. Purified basophils are stimulated with stimuli for 30 min. As degranulation is a fast process, this short time is sufficient. Typical externalization of CD63 and CD203c after stimulation with 1 µg/ml anti-IgE or 1 µM fMLP, in comparison to negative control (Co)-stimulated basophils is depicted in Figure 4. Depending on the potency of stimuli activation, up to nearly 100% of basophils can become activated. If high levels of base level activation are observed, the basophil donor most likely has come into contact with an allergen that they are allergic to, and cells might not be usable for BAT analysis.
Time Considerations
The basic protocol for isolating basophils from peripheral blood will take ∼90 min to perform. The time requirement for Support Protocol 1 to assess basophil purity, yield, and viability is ∼20 min. Support Protocols 2 to 4 will take ∼90 min each.
Acknowledgments
This work was supported by the German Research Foundation DFG with a grant to U.R. RA-1026/3-2 (FOR2690-PruSEARCH Translational Pruritus Research). Further funding was provided by the research pool of the Carl von Ossietzky Universität Oldenburg to U.R. (FP 2019-033) and to M.M.L. and U.R. (FP 2020-053; FP 2023-079). The authors also appreciate the assistance of the cell sorting facility supported by the DFG (ID424196510).
The authors greatly appreciated Alexandra Marten and Insa Kerkhoff for providing excellent technical support, and the team of physician and assistants consisting of Christian Müller, Tara Müller, Tim Schricker, Lea-Sophie Stahl, Lena Fieber, Ingryda Latoschinski, and Katrin Ubber for recruiting the donors and collecting blood samples. The authors also thank Toni Weinhage from Beckman Coulter for sharing his expertise on flow cytometry.
Open access funding enabled and organized by Projekt DEAL.
Author Contributions
Natalie Gray : Conceptualization; data curation; investigation; methodology; validation; visualization; writing original draft. Daniela Wiebe : Investigation; methodology; writing review and editing. Tobias Weihrauch : Investigation; methodology; writing review and editing. Ulrike Raap : Conceptualization; funding acquisition; project administration; resources; supervision; validation; writing review and editing. Maren M. Limberg : Conceptualization; data curation; funding acquisition; investigation; methodology; project administration; supervision; validation; visualization; writing review and editing.
Conflict of Interest
The authors declare no conflict of interest.
Open Research
Data Availability Statement
Data is available from the authors upon reasonable request.
Literature Cited
- Berger, A. (2000). Th1 and Th2 responses: What are they? BMJ , 321, 424. https://doi.org/10.1136/bmj.321.7258.424
- Doña, I., Ariza, A., Fernández, T. D., & Torres, M. J. (2021). Basophil activation test for allergy diagnosis. Journal of Visualized Experiments: JoVE , https://doi.org/10.3791/62600
- Gibbs, B. F., Noll, T., Falcone, F. H., Haas, H., Vollmer, E., Vollrath, I., Wolff, H. H., & Amon, U. (1997). A three-step procedure for the purification of human basophils from buffy coat blood. Inflammation Research , 46, 137–142. https://doi.org/10.1007/s000110050537
- Gray, N., Limberg, M. M., Wiebe, D., Weihrauch, T., Langner, A., Brandt, N., Bräuer, A. U., & Raap, U. (2022). Differential Upregulation and functional activity of S1PR1 in human peripheral blood basophils of atopic patients. International Journal of Molecular Sciences , 23, 16117. https://doi.org/10.3390/ijms232416117
- Iype, J., & Fux, M. (2021). Basophils orchestrating eosinophils' chemotaxis and function in allergic inflammation. Cells , 10, 895. https://doi.org/10.3390/cells10040895
- Kepley, C., Craig, S., & Schwartz, L. (1994). Purification of human basophils by density and size alone. Journal of Immunological Methods , 175, 1–9. https://doi.org/10.1016/0022-1759(94)90326-3
- Li, H., Sim, T. C., & Alam, R. (1996). IL-13 released by and localized in human basophils. Journal of Immunology , 156, 4833–4838.
- Liang, G., Zhou, J., Jiang, L., Wang, W., Wu, Q., Gao, C., Liu, W., Li, S., Feng, S., & Song, Z. (2023). Higher house dust mite-specific IgE levels among chronic spontaneous urticaria patients may implicate higher basophil reactivity. International Archives of Allergy and Immunology , 84(11), 1126–1134. https://doi.org/10.1159/000531966
- Limberg, M. M., Wiebe, D., Gray, N., Weihrauch, T., Bräuer, A. U., Kremer, A. E., Homey, B., & Raap, U. (2023). Functional expression of TRPV1 in human peripheral blood basophils and its regulation in atopic dermatitis. Allergy , 79, 225–228. https://doi.org/10.1111/all.15802
- MacGlashan, D., White, J. M., Huang, S. K., Ono, S. J., Schroeder, J. T., & Lichtenstein, L. M. (1994). Secretion of IL-4 from human basophils. The relationship between IL-4 mRNA and protein in resting and stimulated basophils. Journal of Immunology , 152, 3006–3016.
- Martelletti, P., Adriani, E., Bonini, S., Celestino, D., Lenti, L., Armaleo, C., Di Pastena, A., Misasi, R., & Giacovazzo, M. (1989). Basophil histamine release and leukotriene (LTB4-LTC4) production in cluster headache. Headache , 29, 46–48. https://doi.org/10.1111/j.1526-4610.1989.hed2901046.x
- Miroli, A. A., James, B. M., & Spitz, M. (1986). Single step enrichment of human peripheral blood basophils by Ficoll-Paque centrifugation. Journal of Immunological Methods , 88, 91–96. https://doi.org/10.1016/0022-1759(86)90055-4
- Pösel, C., Möller, K., Fröhlich, W., Schulz, I., Boltze, J., & Wagner, D.-C. (2012). Density gradient centrifugation compromises bone marrow mononuclear cell yield. PLoS ONE , 7, e50293. https://doi.org/10.1371/journal.pone.0050293
- Raap, U., Gehring, M., Kleiner, S., Rüdrich, U., Eiz-Vesper, B., Haas, H., Kapp, A., & Gibbs, B. F. (2017). Human basophils are a source of - and are differentially activated by - IL-31. Clinical and Experimental Allergy , 47, 499–508. https://doi.org/10.1111/cea.12875
- Raghuprasad, P. K. (1982). A rapid simple method of basophil purification by density centrifugation on Percoll. Journal of Immunology , 129, 2128–2133.
- Saini, S. S., MacGlashan, D. W., Sterbinsky, S. A., Togias, A., Adelman, D. C., Lichtenstein, L. M., & Bochner, B. S. (1999). Down-regulation of human basophil IgE and FCεRIα surface densities and mediator release by Anti-IgE-infusions is reversible in vitro and in vivo. Journal of Immunology , 162, 5624–5630. https://doi.org/10.4049/jimmunol.162.9.5624
- Schroeder, J. T., & Bieneman, A. P. (2016). Isolation of human basophils. Current Protocols in Immunology , 112, 7.24.1–7.24.8. https://doi.org/10.1002/0471142735.im0724s112
- Shiono, H., Matsui, T., Okada, T., & Ito, Y. (2016). Single-step enrichment of basophils from human peripheral blood by a novel method using a Percoll density gradient. Journal of Separation Science , 39, 3062–3071. https://doi.org/10.1002/jssc.201600329
- Siracusa, M. C., Kim, B. S., Spergel, J. M., & Artis, D. (2013). Basophils and allergic inflammation. Journal of Allergy and Clinical Immunology , 132, 789–801. quiz 788, https://doi.org/10.1016/j.jaci.2013.07.046
- Stone, K. D., Prussin, C., & Metcalfe, D. D. (2010). IgE, mast cells, basophils, and eosinophils. Journal of Allergy and Clinical Immunology , 125, S73–80. https://doi.org/10.1016/j.jaci.2009.11.017
- Wiebe, D., Limberg, M. M., Gray, N., & Raap, U. (2023). Basophils in pruritic skin diseases. Frontiers in Immunology , 14, 1213138. https://doi.org/10.3389/fimmu.2023.1213138

