DNA extraction from feathers
George Olah
Abstract
This protocol describes a DNA extraction method from feathers collected non-invasively in the wild.
Steps
Collection & Preparation
When collecting in the field, avoid touching the tip of the feathers. If wet, dry the feather on paper towel before storing it.
Place each feather sample in a separate paper envelope, labelling: individual sample ID, collection date, location, species, collector's name, etc.
Store the envelopes with samples in an airtight dry-box with Silica gel crystals at Room temperature.
For each feather sample, prepare and label an
For each feather sample, get:
- a clean A4 paper sheet
- a pair of
- a sterile
.
Isolation
Put on the
Carefully clean the surface of the feather with
Remove a
Large feathers: carefully cut out a window around the blood clot from the superior umbilicus of the feather (this can usually be seen just below the vane).
See details in:
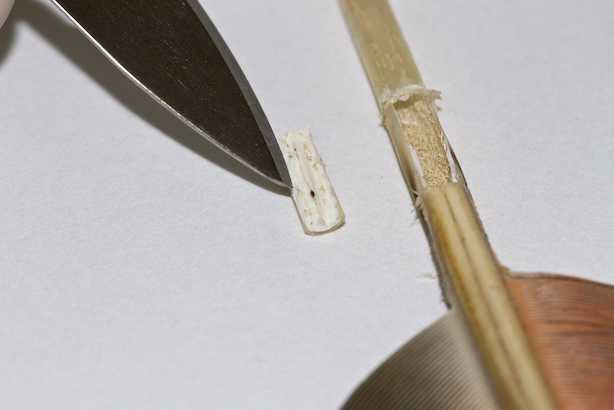
Place the isolated sample into the corresponding Eppendorf tube with the correct sample ID.
Dispose the
Put the remaining part of the feather back to its envelope.
Dispose the A4 paper and the gloves.
and repeat the steps for each feather sample, avoiding cross-contamination.
Negative control: label an empty tube as negative control for the subsequent steps.
Lysis
Add 180µL of
Add 20µL of 20mg/mL to each tube.
Add 10µL of
Vortex each tube for 0h 0m 20s.
Incubate 1h 0m 0s at 56°C on a block heater.
Vortex each tube for 0h 0m 20s.
Incubate at 56°C on a block heater.
Purification
Unpack appropriate number of spin column + collection tubes (according to the number of samples you are extracting) from
Vortex each Eppendorf tube with the samples for 0h 0m 15s.
Spin down briefly.
Add 200µL of
Vortex for 0h 0m 15s.
Spin down briefly.
Incubate 0h 45m 0s at 70°C on a block heater.
Spin down briefly and add 210µL of
Vortex and incubate for 0h 5m 0s at Room temperature.
Spin down briefly and pipette liquid from the Eppendorf tubes to the correspondingly labelled spin columns. Use
Centrifuge 10000rpm.
Discard collection tube with the filtrate and place the spin column in a clean collection tube.
Add 500µL of 10000rpm.
Discard collection tube with the filtrate and place the spin column in a clean collection tube.
Add 500µL of 12000rpm.
Recovery of DNA
Preheat appropriate volume of 100µL x number of samples + a little extra) to 70°C.
Prepare as many
Discard collection tube with the filtrate and place spin column into a clean 1.5 mL Eppendorf tube (with lid cut off).
Add 100µL of preheated 70°C for 0h 5m 0s on a block heater.
Prepare
Centrifuge incubated samples 10000rpm.
Carefully lift the spin column, pipette up the eluted sample from the Eppendorf tube (~100 uL), place back to the spin column into the Eppendorf tube, and pipette the solution back to the center of the filter.
Incubate at 70°C for 0h 1m 0s.
Centrifuge 10000rpm.
Transfer filtrate to labeled
Store screw cap tubes in fridge at 4°C for short-term or in a freezer at -20°C for long-term storage.

