Complementary Tumor Diagnosis by Single Cell–Based Cytogenetics Using Multi-marker Fluorescence In Situ Hybridization (mFISH)
Gero Brockhoff, Gero Brockhoff
Abstract
Multi-color (or multi-marker) fluorescence in situ hybridization (mFISH) is a well-established, valuable, complementary tool for prenatal and pathological (tumor) diagnosis. A variety of chromosomal abnormalities, such as partial or total chromosomal gains, losses, inversions, or translocations, which are considered to cause genetic syndromes, can relatively easily be detected on a cell-by-cell basis. Individual cells either in suspension (e.g., in the form of a cytological specimen derived from body fluids) or within a tissue (e.g., a solid tumor specimen or biopsy) can be quantitatively evaluated with respect to the chromosomal hybridization markers of interest (e.g., a gene or centromeric region) and with due consideration of cellular heterogeneity. FISH is helpful or even essential for the (sub-)classification, stratification, and unambiguous diagnosis of a number of malignant diseases and contributes to treatment decision in many cases. Here, the diagnostic power and limitations of typical FISH and mFISH approaches (except chromosome painting and RNA hybridization) are discussed, with special emphasis on tumor and single-cell diagnostics. Well-established and novel FISH protocols, the latter addressed to accelerate and flexibilize the preparation and hybridization of formalin-fixed and paraffin-embedded tissues, are provided. Moreover, guidelines and molecular aspects important for data interpretation are discussed. Finally, sophisticated multiplexed approaches and those that analyze very rare single-cell events, which are not yet implemented in diagnostic procedures, will be touched upon. © 2023 The Authors. Current Protocols published by Wiley Periodicals LLC.
This article was corrected on 21 December 2023. See the end of the full text for details.
Basic Protocol 1 : (m)FISH applied to formaldehyde-fixed paraffin-embedded tissues
Basic Protocol 2 : (m)FISH applied to cytological specimens
INTRODUCTION
General Considerations for “Cytogenetics” and “mFISH”
The term “cytogenetics” (in the narrower sense) refers to the analysis of chromosomes and related abnormalities that are known to cause genetic syndromes. Cytogenetic analysis is very crucial in the diagnosis of prenatal chromosomal aberrations, infectious diseases, and oncologic and hematologic disorders. Methodologically, cytogenetics comprises a number of techniques with different validity, among which are (array-based) comparative genomic hybridization (a/CGH) (see Current Protocols article: Bayani & Squire, 2005), “G-banding,” and chromogenic in situ hybridization (ISH) (see Current Protocols article: Bayani & Squire, 2004b) or fluorescence ISH (FISH) (see Current Protocols article: Bayani & Squire, 2004a; Lee et al., 2001).
The term “mFISH,” or multi-color (or multi-marker) FISH, is frequently used to refer to so-called “chromosome painting,” which sometimes is alternatively named spectral karyotyping (SKY) or COmbined Binary RAtio labeling-FISH (COBRA-FISH (Liehr et al., 2006; Liehr et al., 2017). By the “painting procedure,” the whole chromosome (or larger segments of it) becomes hybridized, which results in DNA-specific chromosomal color banding to evaluate the number, extent, and location of chromosomal subregions (Langer et al., 2004). The approach even allows “painting” each of the 24 different human chromosomes with different colors (i.e., fluorochromes) at the same time (Azofeifa et al., 2000), but in clinico-pathological diagnostics, it plays just a minor role, if any. This sort of mFISH is discussed elsewhere and is not within the scope of this article.
Instead, this article refers to mFISH in the sense of the simultaneous use of multiple hybridization probes that are suitable to quantify region-specific copy numbers and to identify intra- and inter-chromosomal locations of specific but small (a couple of hundred kilobases) chromosomal regions. In this sense, the cytogenetic procedure refers to the detection and visualization of known and precisely defined chromosomal regions by the sequence-complementary annealing of fluorochrome-labeled nucleic acid probes. The hybridization is applied to permeabilized but morphologically preserved cells or partially digested tissues without extensive cell disruption. Thus, the detection of chromosomal regions of interest occurs in native cellular and/or tissue locations, i.e., in situ. Diagnostic FISH is inherently a quantitative approach inasmuch as DNA hybridization spots are countable and their relative position (to each other) is assessable and recordable. In this way, FISH enables the uncovering of genetic alterations at the chromosomal level in preserved and immobilized single cells or tissue specimens.
A prenatal cytogenetic (F)ISH diagnosis is typically performed based on chorionic villus sampling or amniocentesis during pregnancy (Pergament et al., 2000; Tepperberg et al., 2001). This approach aims to determine if a fetus is at risk for, e.g., common aneuploidies/polysomies (syndromes caused by having extra or missing chromosomes) or syndromes caused by structural abnormalities (like unbalanced translocations or inversions) or to determine if there is extra or missing genetic material (i.e., total or partial gains or losses of chromosomes). Clinically, chromosome abnormalities account for a large proportion of cases involving individuals referred with congenital malformations, developmental delay, intellectual disability, or infertility; women with gonadal dysgenesis or spontaneous abortions; and couples with repeated spontaneous miscarriages.
This article, however, focuses on ISH and (m)FISH applications in oncology. Similar to prenatal analyses in tumor diagnostics, (F)ISH tracks down to chromosomal aberrations and abnormalities and thereby complements pathological diagnosis, supports classification and stratification of malignant diseases, and provides prognostic (and sometimes predictive) information. Today, the complete diagnosis and treatment of several solid cancers (e.g., breast, lung, bladder, and gastric cancer) and a number of malignancies with mesenchymal origin (e.g., leukemias and lymphomas, rhabdomyosarcoma, Ewing sarcoma, liposarcoma, synovial sarcoma) heavily depend on cytogenetic analysis of specific chromosomal aberrations that are consistently observed in malignant cells of these diseases. Hence, region-specific marker hybridizations are often helpful or even essential in planning individual treatment regimens and in monitoring the status of a malignant disease (Bjerregaard et al., 2018; Zhang et al., 2014; Zhou et al., 2016).
FISH can be applied to metaphase-spread chromosome preparations and to interphase cell nuclei. The latter approach is commonly applied to fresh (i.e., non-fixed) tissues, to cytology preparations, or to paraffin-embedded specimens. Which approach is preferably applied depends on the type of study requested. The significant advantage of FISH (i.e., the use of fluorophore-labeled probes) over ISH (i.e., the use of chromogenic labeled probes) is the ability to apply and visualize more than two probes simultaneously, which results in a multi-color (i.e., multi-marker) hybridization (mFISH). In this way, multiple regions located on the same chromosomes or on different chromosomes can be addressed and analyzed at the same time (Denzinger et al., 2007; Miyake et al., 2023; Pothos et al., 2008; Schwarz et al., 2004; Schwarz et al., 2008). Additionally, individual markers can be visualized and assessed separately or simultaneously by using microscopic single-color or dual- or even three-color filters. The use of multiple probes enhances the diagnostic power in many cases and enables one to address diagnostic questions that could often not be answered by using only one or two marker probes.
With respect to cancer treatment, FISH not only identifies genetic or chromosomal anomalies that drive carcinogenesis and tumor progression but also identifies those abnormalities that result in the (over-)production of (potential) therapeutic targets. Examples are (i) the gene fusion product BCR/ABL1 (i.e., the Philadelphia chromosome), which is generated by a t(9;22)(q34.1;q11.2) translocation and frequently found in chronic myeloid leukemias (Shao et al., 2015; Virgili & Nacheva, 2010), and (ii) the HER2/ErbB2 receptor tyrosine kinase gene, which is amplified in ∼20% of all breast (Slamon et al., 2001) and gastric (He et al., 2013) cancers. Thus, patients who are eligible for target-specific treatments can be identified by cytogenetic analyses.
Principles of Hybridization Probes and Procedure
A huge number of hybridization probes used in research and diagnostics are commercially available. Most of them target either repetitive sequences (which are found, for instance, in the centromere (cen) and satellite regions of a chromosome) or highly specific (gene) regions that are unique within the genome. Basic steps of (F)ISH include thermic DNA denaturation in situ (i.e., at the original nuclear location without prior extraction of nucleic acid), which is typically (but not necessarily; see below) performed on a slide. As a result, the DNA becomes single stranded. Afterward, sequence-specific DNA probes are added to the specimen and hybridized to the target sequences under stringent conditions (i.e., annealing of probes under renaturating conditions). This means that the hybridization conditions are adjusted in such a way that, instead of the original complementary DNA strands, the probes preferentially bind to the target region. This includes, for example, the probe concentration (excess) and the temperature. As a result, fluorescent hybridization spots are generated at specific chromosomal loci. For the later visualization of sequence-specific hybridizations, the DNA probes are labeled with a chromogen or a fluorochrome. After the hybridization procedure is complete, FISH specimens can be conserved by slide sealing, and finally, the fluorescent hybridization spots can be microscopically visualized. Hybridization results can be manually or semi-automatcially (see below) analyzed. The object of the final analysis is basically the quantification and (dependent on the diagnostic question and probe design) the location of hybridization signals (i.e., colored fluorescent spots) on a cell-by-cell basis (more specifically, nucleus by nucleus).
Cytogenetic Evaluation of Single Cells by Metaphase mFISH
Metaphase FISH studies are designed to detect changes (i.e., microdeletions and/or microduplications) associated with specific phenotypic findings and require specific and focused clinical questions to be addressed via appropriate DNA probe selection. Such an approach provides an overview of the entirety of the chromosomal aberrations in a single tumor cell. However, metaphase FISH testing is limited to those DNA probes that are currently available and validated for clinical use. Genetic changes that are detected in metaphase chromosome preparations are limited to position and copy number changes, primarily losses (deletions) and, in some instances, gains (duplications) of specific chromosomal regions. Nevertheless, the repositioning of DNA probes from their normal sites can be determined in metaphase cell preparations (i.e., translocations, insertions) (Ohno et al., 2022). However, these types of studies do not rule out other forms of genetic abnormalities, which may include low-level or tissue-specific mosaicisms and/or other forms of molecular alterations (i.e., single-base pair mutations or uniparental disomies, among others).
Methodically, the pretreatment of specimens with colchicine or colcemid results in the destruction of the mitotic spindle apparatus and thereby causes the accumulation of dividing (mitotic) cells in metaphase of the cell cycle. Hypotonic solutions cause swelling of the cell membrane and the dispersion of chromosomes so that the enumeration and further analysis of the chromosome structure in individual (human) cells become possible. The procedure of metaphase FISH has been described in detail elsewhere (MacLeod et al., 2017).
Cytogenetic Evaluation of Single Cells by Interphase mFISH
Interphase FISH is done without prior cell cycle synchronization. It provides information regarding the number and/or rearrangement of specific chromosomal DNA regions. Even though other structural inferences are not possible, interphase FISH is being frequently used in clinico-pathological diagnostics. (m)FISH testing based on interphase specimens is typically applied to identify the presence, absence, relative positioning, and/or frequency (i.e., copy number) of specific DNA segments by (fluorescence) microscopy. It plays a major role in pathological tumor diagnostics.
The quantification of hybridized marker probes is commonly done by microscopic visualization of hybridization spots and manual counting of those spots on a single-cell level. Alternatively, the identification of cells (within a tissue) and the FISH signal counting can be performed (semi-)automatically using sophisticated software and imaging systems (Knudson et al., 2007; Martinez et al., 2013; Narath et al., 2004; Nishimura et al., 2016; Smith et al., 2010; Zwaenepoel et al., 2015). Automated cell imaging and analyses have been successfully applied using systems developed and distributed, for example, by BioView (www.bioview.com) or Metasystems (www.metasystems-international.com). Appropriate software enables automated identification of single cell nuclei and hybridization spots therein. However, a manual inspection of software-generated results is advisable and often even necessary, which can be done by inspection of the specimens itself or of the related image galleries.
Applying interphase FISH to tissue specimens enables the evaluation of single cells within the context of the tissue architecture. Thus, individual tumor cells can be cytogenetically assessed with due consideration of well-established pathological parameters, e.g., grade of tissue differentiation, scirrhous tumor growth, cell morphology, and tumor heterogeneity (Daniely et al., 2007). Often 20 or more cells (up to 50) are evaluated individually within a tissue. In cases where more than one marker probe is applied simultaneously, the counting of all marker spots takes place nucleus by nucleus rather than marker by marker. This counting procedure ensures capture of not only average hybridization values for individual markers for a given number of cells (or rather, cell nuclei) but also particularly the marker values for individually evaluated cells. As a result, not only the average cytogenetic status of a tissue but also the status of individual cells is assessed. If two or more marker regions, for example, cen- and gene-specific regions, are hybridized at the same time, it is advisable to independently quantify their individual frequencies and to interrelate their frequencies by calculating marker ratios (see the following section for more details).
HER2 Single-Marker and HER2/cen17 Dual-Marker FISH in Breast Cancer (in Combination with HER2 Immunofluorescence)
One of the most prominent examples of region-specific, dual-marker (cen and gene regions) FISH is probably the HER2/cen17 hybridization approach, which plays a pivotal role in the diagnosis of breast (and gastric) cancer. The clinical importance of HER2 gene amplification and corresponding receptor overexpression has been known since the mid-1980s (Slamon et al., 1987). About 20% of all breast cancers are considered diagnostically HER2 positive (Slamon et al., 2001). The prognosis for HER2-positive breast cancer patients is rather poor; however, a specific therapeutic targeting HER2 (e.g., trastuzumab) significantly improves the outcome and elevates the prognosis for HER2-positive breast cancer patients up to the level of that of HER2-negative breast cancer patients without such a treatment (Dawood et al., 2010). Thus, HER2 testing has great clinical impact and has become mandatory for the diagnosis of invasively growing breast cancer.
It has been repeatedly reported that HER2 protein overexpression is predominantly, if not exclusively, caused by an increased HER2 gene copy number (McCormick et al., 2002; Li-Ning-T et al., 2005; Lottner et al., 2005). For this reason, the concordance between HER2 gene copy number (assessed, for example, by PCR or FISH) and HER2 protein overexpression (immunohistochemically scored as 3+) is very high (Bilous et al., 2006; Koudelakova et al., 2015; Li-Ning-T et al., 2005; Lottner et al., 2005; Sassen et al., 2008). Guidelines for FISH processing, testing, and data interpretation were released by the American Society of Clinical Oncology/College of American Pathologists (ASCO/CAP) years ago (Wolff et al., 2007) and have been updated since, e.g., in 2013 and 2018 (Wolff et al., 2013; Wolff et al., 2018). However, multiannual experience with HER2 testing in breast cancer made it apparent that the immunohistochemical approach is subject to certain variabilities for intra- and inter-laboratory technical reasons and, to some extent, due to variation in observer-dependent data interpretation (Green & Zynger, 2015). In contrast, the preparation of FISH specimens turned out to be more robust, and the spot counting of FISH specimens occurred more objectively because it is basically a user-independent and particularly quantitative approach. Consequently, the FISH approach evolved as a diagnostic “gold standard” for HER2 testing.
Due to enhanced diagnostic power, dual-marker (cen and gene locus) FISH is certainly more frequently applied than the hybridization of a HER2 probe only. The single cell–based evaluation allows the absolute quantification of cen and gene copy numbers of individual cells, enables the calculation of average values for individual specimens, and facilitates the calculation of marker ratios (the number of HER2 signals divided by the number of cen hybridization spots). All these data have different but complementary diagnostic value, according to the following interpretation (Wolff et al., 2013; Wolff et al., 2018).
Cen probes target chromosome-specific repetitive sequences. Because the non-coding centromeric region of a chromosome is assumed to appear solely (i.e., independently from other chromosome regions) not altered (i.e., deleted or amplified), the cen copy number is supposed to reflect the number of the respective chromosome. On this understanding, the hybridization of the cen region allows the differentiation of disomy, polysomy, and monosomy. Even a total loss of both copies of a particular chromosome is possible.
Unlike hybridization of a cen region, the sequence-specific hybridization of a gene region is considered to truly reflect the gene frequency within a given cell nucleus. Because the “normal” (i.e., unaltered) human somatic cells are generally disomic (i.e., all chromosomes are present twice in a nucleus), fewer than two hybridization signals are interpreted as a gene loss (i.e., deletion), whereas more than two signals are considered to reflect a gene copy gain. A gain might be due to either a real gene amplification or polysomy of the respective chromosome (which also results in an increased gene dose) or a combination of both. Example images of different HER2/cen17 hybridization results are shown in Figure 1.
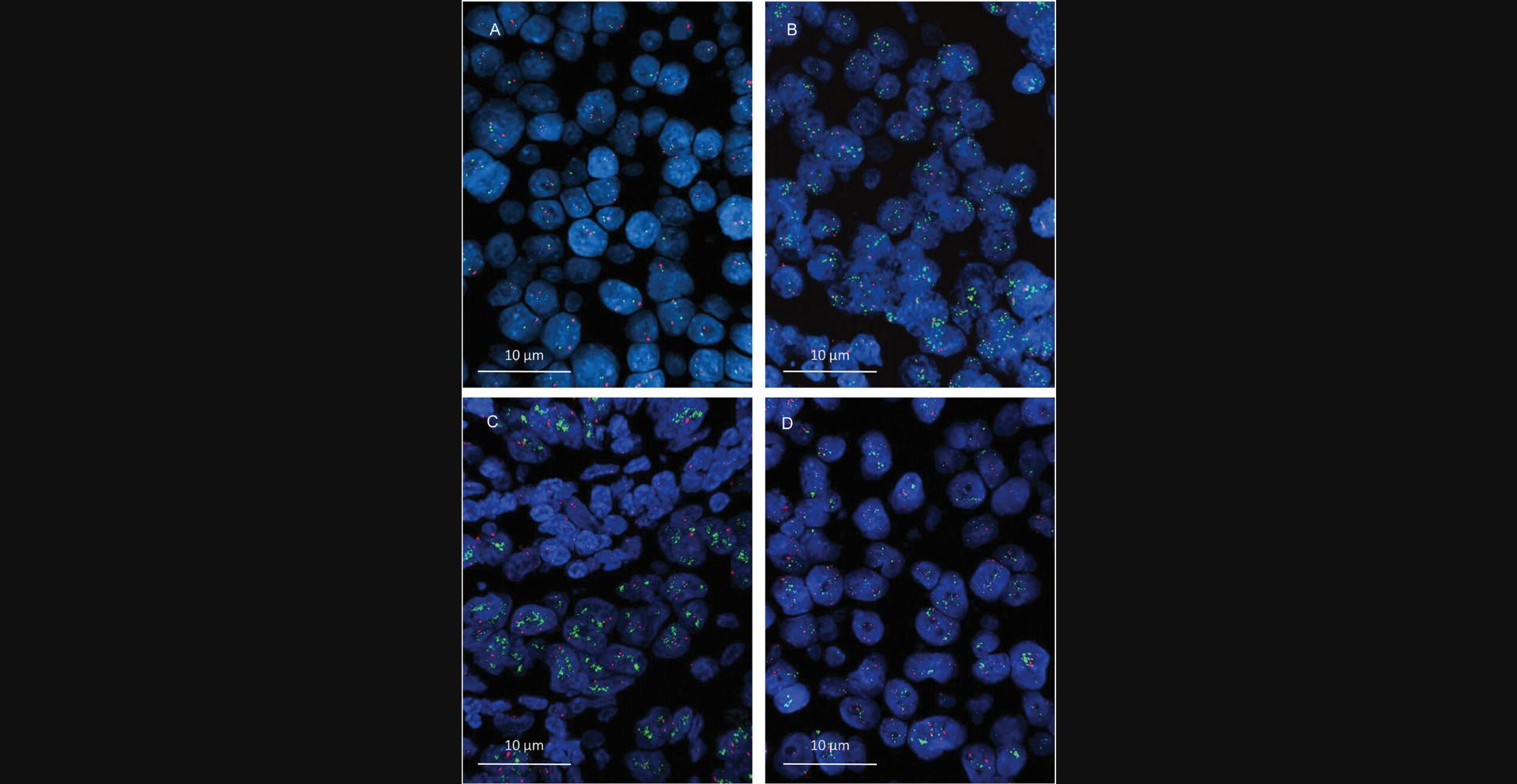
In contrast to two-marker HER2/cen17 FISH diagnostics, probing the HER2 gene region alone is of course also possible but is less informative because it does not provide any clue about the origination of the visualized increased HER2 gene dose. More specifically, a real HER2 gene amplification or the presence of (putative) polysomy 17 cannot be differentiated. This distinction is, however, relevant, because breast cancers with an enhanced HER2 gene dose, which is due to either real gene amplification or a multiplied chromosome 17, are considered to represent different prognostic entities that might be reasonably differently treated (Perez et al., 2010; Hanna et al., 2014).
It must be mentioned, however, that the cen region of a chromosome does not necessarily work as an integer surrogate for the chromosome number, especially with respect to chromosome 17. Instead, an increased cen17 copy number has been demonstrated to occur not rarely in the absence of true polysomy as a result of multiplication within an amplicon (Marchiò et al., 2009; Koudelakova et al., 2016; Donaldson et al., 2017; Holzschuh et al., 2017), which comprises several chromosomal (coding and not-coding) regions in close proximity. The differentiation of an amplified cen region versus true polysomy would, however, require additional genomic analysis, for example, with array-based comparative genomic hybridization (Holzschuh et al., 2017), multiplex ligation-dependent probe amplification (Pazhoomand et al., 2013), or quantitative real-time PCR (Koudelakova et al., 2015). Nonetheless, the clinical relevance of mixed genotypes (true and putative polysomy) within single cells or individual tumor tissues has not been definitively clarified.
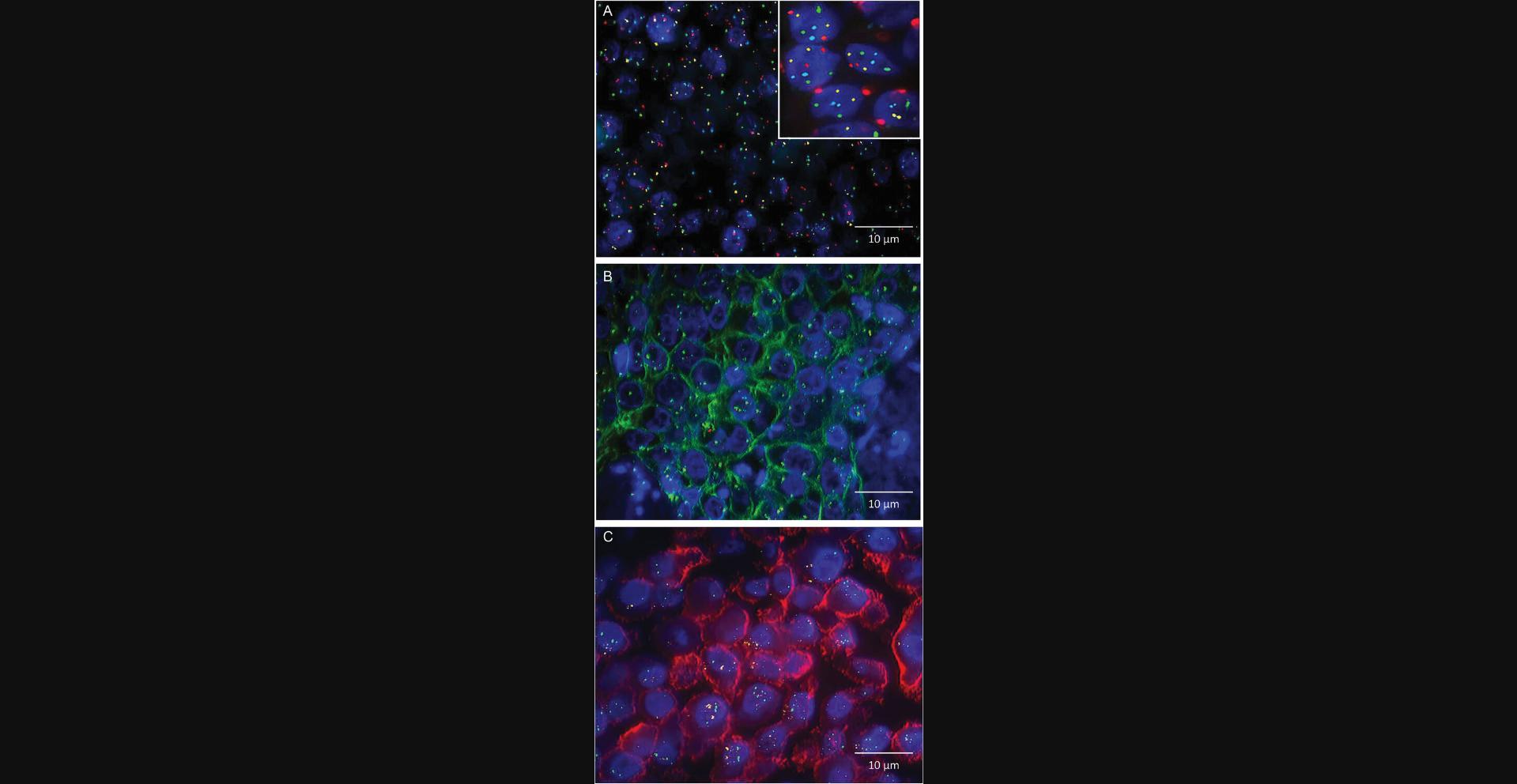
As mentioned above, a big advantage of subjecting tissues to interphase FISH is the feasibility of assessing a number of single cells in the context of a variety of diagnostically relevant parameters (e.g., cellular heterogeneity, cell morphology, growth characteristics of different tissue areas). Thus, extending the number of parameters assessed in single cells within a tissue enhances the analytical power. Accordingly, a diagnostic four-color approach that combines two-marker hybridization with fluorescence-based immunohistochemistry (FIHC) on formaldehyde-fixed paraffin-embedded (FFPE) breast cancer specimens was established some time ago (Lottner et al., 2005). The multiplex single-step analysis enables quantification of the HER2 gene copy number, assessment of (putative) polysomy 17, and semi-quantitative scoring of the HER2 protein expression of single cells. The approach has been validated on 215 primary breast carcinomas, showing an excellent overall correlation between an FIHC score and the corresponding HER2 gene copy number. The potential advantage is that FISH and FIHC can be applied simultaneously (instead of sequentially) to identical tissue specimens so that potential discrepancies between gene dose and protein expression can be uncovered. Overall, the combined FIHC/FISH application demonstrates the power of fluorescence-based (semi-)quantification of multiple cellular parameters, which can be adapted to the analysis of other genomic and protein markers. Examples of multiprobe/multiplex FISH analyses are given in Figure 2.
Detection of Chromosomal Rearrangements by FISH
Beyond marker enumeration, interphase FISH is diagnostically applied to identify chromosomal marker rearrangements (i.e., translocations). Numerous solid cancers, sarcomas, and leukemic diseases contain malignant cells in which total or partial gene regions underwent intra- or inter-chromosomal translocation. A mutual translocation can cause either an enhanced or a silenced control of gene transcription or can result in the synthesis of functional proteins with new properties that drive malignant cell growth. This sort of chromosomal rearrangement is known, for example, in lung cancer (region 2p23.1-p23.2 harboring the ALK gene), in alveolar rhabdomyosarcomas (region 13q14.11 harboring the FOXO1 forkhead box O1 gene), and in chronic myeloid leukemia (regions 9q34.12 and 22q11.23 harboring the ABL1 and the BCR gene regions). The BCR/ABL1 fusion product, which has been named the “Philadelphia chromosome,” results in the generation of a chimeric enzyme with enhanced tyrosine kinase activity that in turn stimulates mitogenic downstream signaling. Note that the list of malignant diseases with potential chromosomal rearrangements in the form of translocations significantly exceeds the number of selectively given examples here and comprises a huge variety of entities.
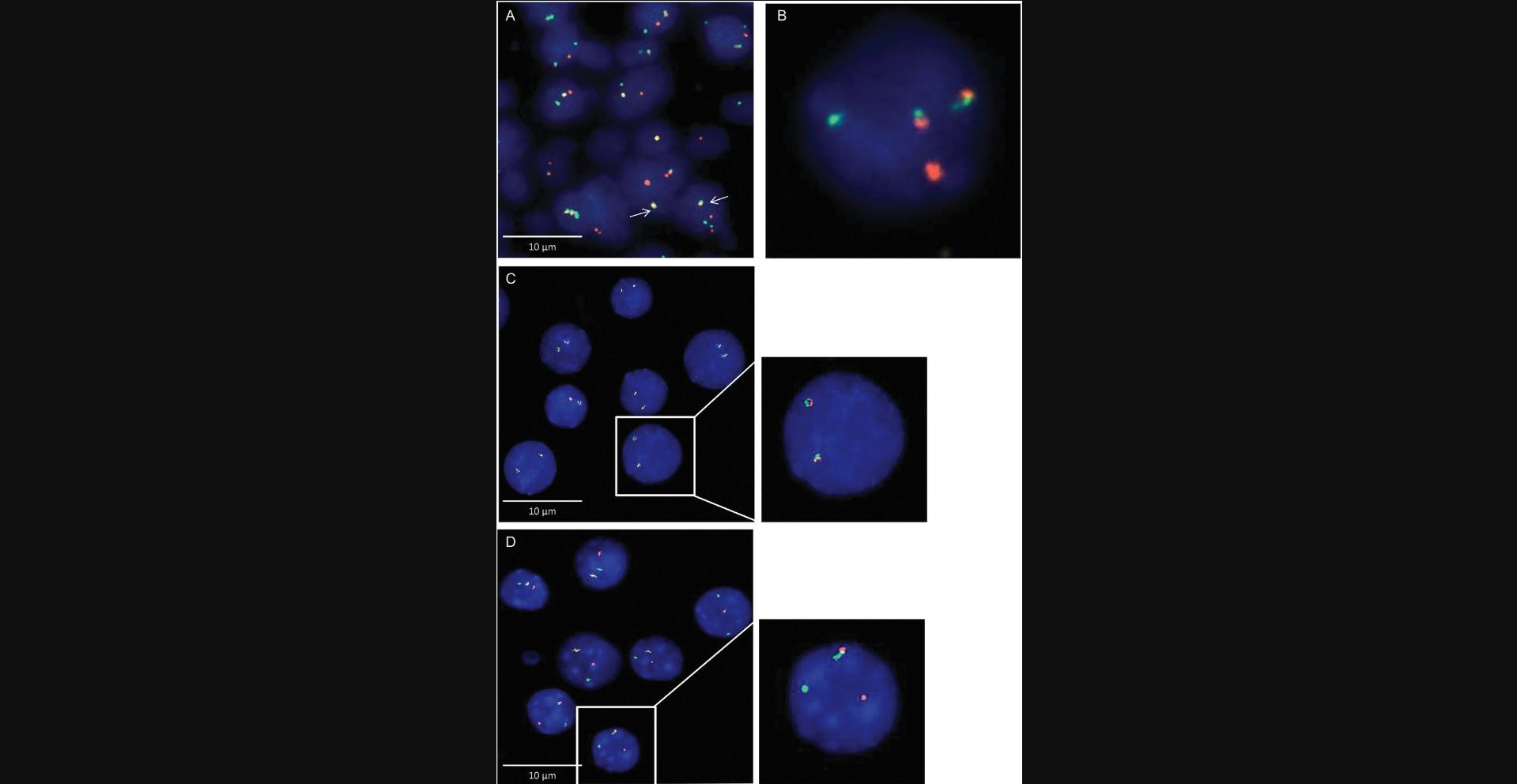
Chromosomal translocations can be detected in a somewhat sophisticated way using so-called “fusion” or “break-apart” probes. The idea is to hybridize two specific chromosomal loci that are known to participate in rearrangements and relocation far from each other. If the translocation generates a larger distance between the two regions, they “break apart.” Conversely, a fusion probe indicates a translocation that results in two closely located regions. Example images are given in Figure 3.
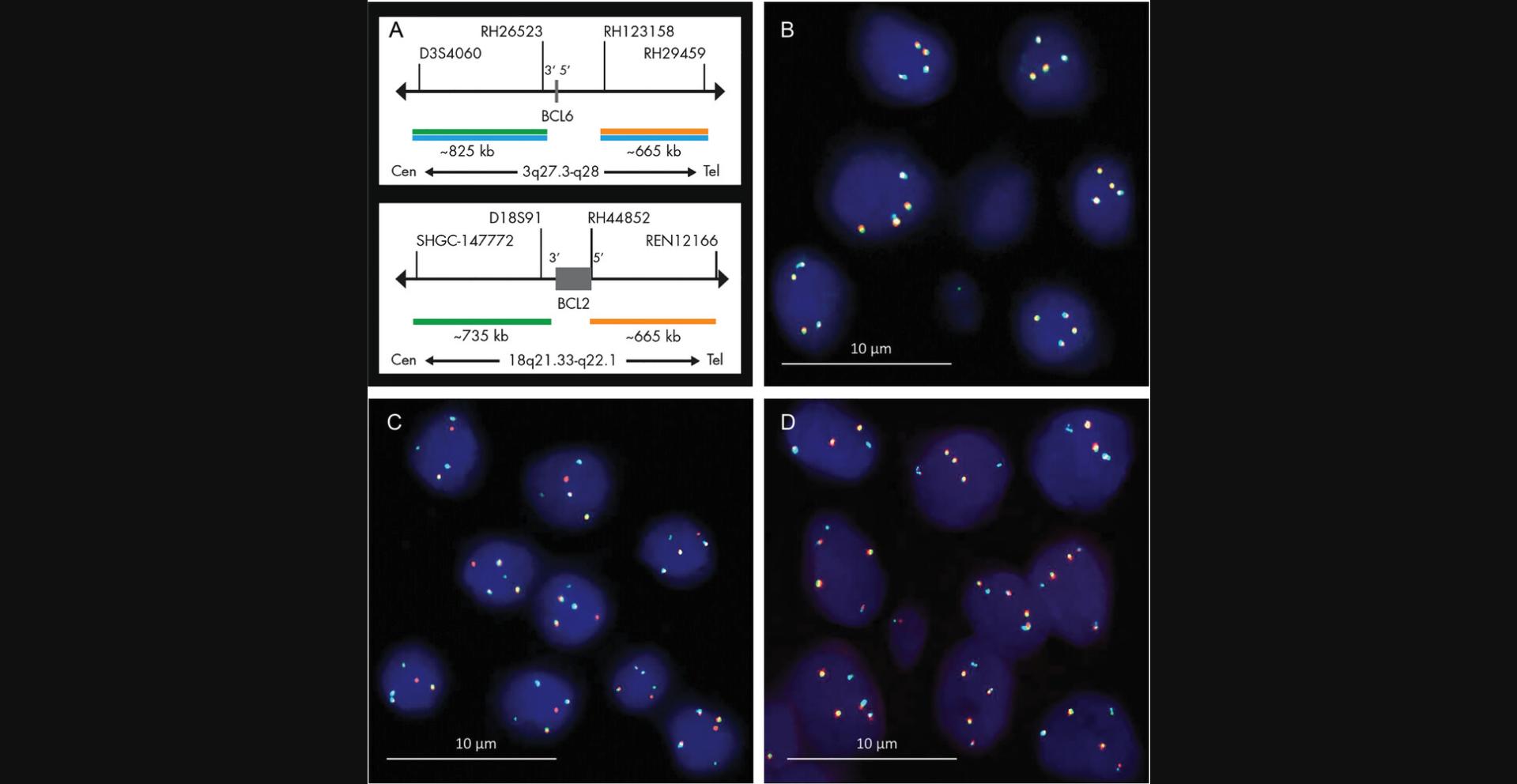
A recent advancement of “conventional” break-apart and fusion probes, with which the translocation of two definite chromosomal loci can be detected, is the development of tricolor probes that allow determination of whether or not certain markers are involved in an observed chromosomal rearrangement. For example, rearrangements that involve either the B-cell lymphoma gene locus 2 or 6 (i.e., bcl2 or bcl6), which are frequently found in various non-Hodgkin lymphomas, can be detected and distinguished by a three-color/five-marker hybridization procedure. Bcl2 encodes a mitochondrial membrane protein that regulates apoptosis and is expressed in B cells. Bcl6 encodes a protein that acts as a transcriptional repressor involved in the regulation of lymphoid development and function. As illustrated in Figure 4A, the so-called bcl2/bcl6 “DistinguISHTM Probe” is a mixture of five direct-labeled probes hybridizing to the 18q21.33-q22.1 and 3q27.3-q28 loci. The green fluorescent probes hybridize proximally to the bcl2 and bcl6 breakpoint regions, whereas the orange fluorescent probes hybridize distally to the bcl2 and bcl6 breakpoint loci. However, the blue fluorescent probe hybridizes both distally and proximally to the bcl6 breakpoint region. In an interphase nucleus without bcl2 or bcl6 rearrangements, two bcl2-specific green/orange fusion signals and two bcl6-specific green/orange/blue fusion signals can be found (Fig. 4B). A bcl2 rearrangement is indicated by one separate green and one separate orange signal, both not co-localizing with blue signals (Fig. 4C). In contrast, a bcl6 rearrangement is indicated by one separate green and one separate orange signal, both co-localizing with blue signals (Fig. 4D). The differentiation of locus-specific rearrangement in non-Hodgkin lymphomas (and other malignancies) has prognostic and therapeutic relevance (Salam et al., 2020; Nakamura et al., 2022).
Another example of advanced ISH that addresses multiple chromosomal regions is the differentiation of ALK inversions and translocations on chromosome 2 in lung cancer, which occur either with or without the involvement of the EML locus. Moreover, an ALK-EML translocation can be discriminated from those that involve the TFG or KIF5B locus, which might have a prognostic and even predictive impact. Another example is the gene fusion of the PDGGRA1 region on chromosome 4. PDGGRA1 fusion might involve a variety of partner regions/genes, among them FIP1L1, BCR, ETV6, KIF5B, STRN, and CDK5RAP2, that can be diagnostically differentiated. The concept of differentiated loci analysis upon rearrangement is illustrated in Figure 5.
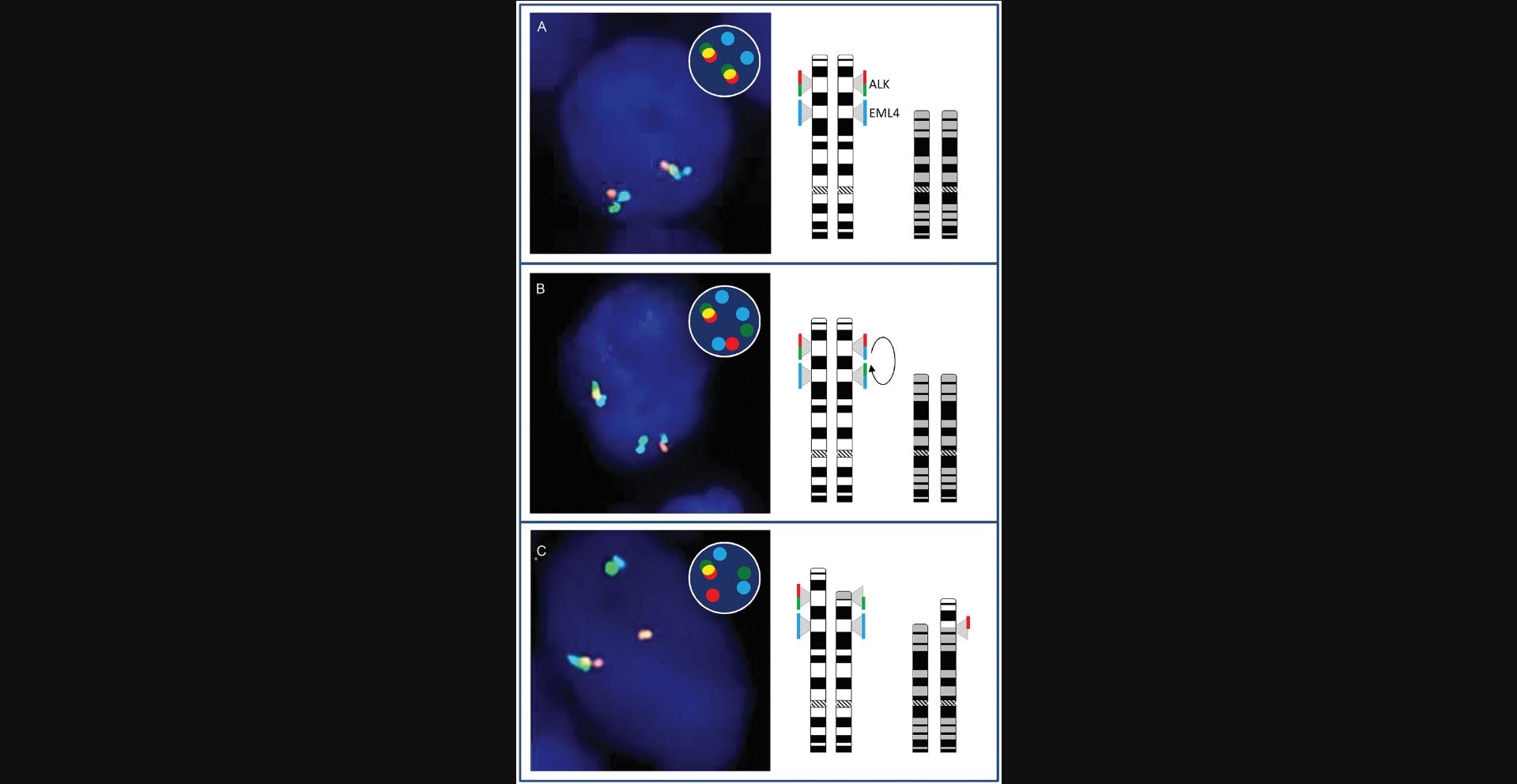
Examples of Advanced (But Diagnostically Not Yet Implemented) Interphase mFISH Applied to Tumor Tissues and Isolated Single Cells
Multi-marker mFISH on rare circulating and disseminated tumor cells
Interphase mFISH is not only being successfully applied to single cells within a tissue; indeed, it has been demonstrated to be helpful in the characterization of rare cells upon isolation from the peripheral blood, an application that is very helpful for the understanding of the process of tumor cell dissemination and metastasis.
Cells that are able to detach from a primary tumor can enter the body periphery as so-called circulating tumor cells (CTCs). Those cells are considered to have (or to develop) the ability to further disseminate into the bone marrow niche, from where they sooner or later form secondary local or distal metastases. Thus, CTCs are gaining the status of a surrogate marker for systemic disease. Recent technological advancements in rare-cell analysis have facilitated the detection of CTCs in the peripheral blood of cancer patients. Numerous clinical studies involving the enumeration of CTCs in cancer patients have unequivocally demonstrated the prognostic value of these cells. However, the outcome of disease is not only determined by the frequency of CTCs but also predominantly by their (inherent or obtained) stemness and resistance to tumor treatment. Hence, the molecular characterization of these very rare cells is of particular interest.
The feasibility of single-cell mFISH applied to CTCs has been demonstrated by Swenninhuis et al. (2009). Upon EpCAM expression–based CTC enrichment using the CellTracks AutoPrep System (Veridex) CTCs were isolated and transferred into a special cartridge, namely a MagNestTM cell presentation device (Veridex). Validated four marker–based identification, fixation, and immobilization of CTCs on the cartridge glass enabled the subsequent application of chromosome-specific (i.e., cen-specific) and fluorescence-labeled hybridization probes. The proof-of-principle study by Swennenhuis et al. revealed a high degree of aneuploidy in CTCs in hormone-refractory prostate cancer patients. More specifically, an enormous heterogeneity with respect to polysomy 1, 7, 8, and 17, both between CTCs derived from one sample and between CTCs derived from different patients, has been observed.
Ignatiadis et al. (2011) used a similar approach to assess the HER2 status of different breast cancer cells with which peripheral blood was spiked. They successfully identified and selected the spiked cells using the CellSearch System (Veridex) and then subjected the tumor cells to HER2 scoring by immunochemistry and FISH. Krishnamurthy et al. (2013) and later Frithiof et al. (2016) extended this proof-of-principle study and demonstrated a profound discordance of HER2 gene amplification in CTCs and disseminated tumor cells (DTCs) compared to primary (breast) tumor cells (PTCs). Using a microfluidic platform for the isolation and enrichment of CTCs and DTCs and based on a simultaneous evaluation of cytokeratin-positive, CD45-negative, and DNA-containing events, a scoring of cen17 and HER2 copy numbers was performed, with 15% discordance between CTCs and PTCs and 28% discordance between DTCs and PTCs. Overall, the study revealed different degrees of genetic instability in PTCs, CTCs, and DTCs derived from individual patients. Equally important, HER2-positive CTCs and DTCs were found in patients with early-stage disease, which suggests that clonal selection or acquisition due to genetic instability can be encountered very early in the course of the disease. Not long ago, Hoffmann et al. (2018) demonstrated that, first, DTCs of patients with esophageal cancer show heterogeneity and, second, only those DTCs with specific chromosomal alterations (i.e., HER2 gene amplifications) determine the patient's survival. These findings exemplify that (m)FISH applied to rare single cells can contribute to extending understanding of tumor progression and can even reveal prognostic and predictive information.
Flow-FISH
The application of FISH is not restricted to embedded tissues or immobilized cells. Specific DNA sequences can also be hybridized in single cells in suspension (FISH-IS). By using fluorochrome-labeled probes, the target cells can be quantitatively analyzed by flow cytometry, which represents a technique that is suited for the quantitative assessment of high numbers of single cells (in suspension) within a relatively short period of time (thousands of cells per second).
A combined Flow-FISH approach has been successfully applied for the analysis and quantification of the lengths and heterogeneity of chromosomal ends in single cells. The distal chromosomal regions, called telomeres, are known to play a fundamental role in cellular senescence and carcinogenesis. On the one hand, a gradual loss of telomeric DNA sequences, which results in a loss of essential (coding and non-coding) chromosomal regions, contributes to cellular aging and finally cell death. Thus, short telomeres represent a risk factor in age-related diseases. On the other hand, anomalous maintenance of telomeres in somatic cells contributes to neoplastic transformation. Hence, a relative or even absolute assessment of telomere lengths in single cells might deliver additional diagnostic information.
A comprehensive protocol dedicated to the measurement of the average length of telomeres by Flow-FISH has been published by Baerlocher et al. (2006). The protocol is based on the use of labeled peptide nucleic acid (PNA) as a telomere-specific probe in combination with a nonspecific DNA-intercalating dye (e.g., LDS-751). Under low-ionic conditions, PNA (but not single-stranded DNA) can anneal to complementary telomeric single-stranded DNA sequences, whereby the telomere lengths can be measured.
Moreover, a procedure for FISH-IS that enables the enumeration of chromosomes in single cells has been described previously (Maguire et al., 2016). Another application of Flow-FISH has been reported by Maiti (2016), who presented a protocol that facilitates the evaluation of the effect of expansion conditions on natural killer (NK) cells in vitro. Because telomere length is known to decrease in adult human NK cells and is associated with proliferation-induced senescence, Flow-FISH-based assessment of telomeres might help to determine the most efficient culture conditions. Additionally, Flow-FISH has been successfully applied for the analysis of viral infection of eukaryotic cells (Manaresi et al., 2015). Finally, this approach has even been reported as an applicable method for the diagnosis of BCR-ABL1 transcripts in leukemic cells (Ranjbaran et al., 2016). It has been impressively demonstrated that Flow-FISH takes advantage of the high specificity of FISH and high sensitivity of flow cytometry.
Last, but not least, it is worth noting a rather sophisticated approach that takes advantage of the combination of FISH-IS, flow cytometry, and imaging of single cells. Using the imaging flow system ImageStream® (Amnis) enables the assessment of thousands of cells in suspension within a short amount of time and their simultaneous visualization. The technique has been applied to FISH-IS-prepared cells on which automated counting and localization of FISH spots has been performed. The application includes the detection of polysomy/aneuploidy, gene amplification and translocation, and, remarkably, rare cells associated with residual disease. Overall, the high throughput of cells analyzed by FISH-IS improves the detection of rare events compared to conventional (slide-based) FISH (Minderman et al., 2012; Maguire et al., 2016).
Non-fluorescent ISH
The power of FISH is the possibility of multi-marker analysis. However, FISH requires a fluorescence microscope for signal visualization. Alternatively ISH, that refers to chromogenic in-situ hybridization can be applied. In contrast to FISH probes, ISH probes are not labeled with chromophores that appear in specific color. A number of product kits are commercially available designed for the detection of aneuploidies, gene copy numbers (gains and losses), and translocations using brightfield microscopy. Like fluorochrome labeled hybridization probes the chromogenic probes can be applied to FFPE tissues, cytological smears, and metaphase chromosome spreads. Evaluating ISH specimens makes it easier to correlate the hybridization result with the cell morphology and tissue architecture. Cytogenetic dual marker HER2/cen17 testing using fluorescent in comparison with chromogenic hybridization probes has been repeatedly evaluated and a great concordance has been mostly reported (Hauser-Kronberger, & Dandachi, 2004; Li-Ning-T et al., 2005; Loring et al., 2005; Pothos et al., 2008; Kato et al., 2010). However, ISH commonly facilitates the hybridization of not more than two markers simultaneously. Visualization requires probe labeling, for example, with digoxigenin which can be detected by an enzyme (e.g., with horse radish peroxidase) conjugated anti-digoxigenin antibody. Alternatively, and analogously to immunohistochemistry, the binding of a non-conjugated anti-digoxigenin antibody can be visualized by an enzyme-conjugated second-step antibody. Finally, the enzymatic reaction of DAB leads to the formation of strong permanent brown color that can be inspected by light microscopy. For probing two markers simultaneously digoxigenin and dinitrophenyl labeled probe kits are available. The application of a peroxidase/phosphatase-linked antibody cocktail for the detection of digoxigenin/dinitrophenyl enables two-color (brown, red) staining of probe hybridization. A typical application of ISH in tissue specimens is for the detection and discrimination of cellular infections with human pathogen viruses [e.g., human papilloma virus (HPV), cytomegalovirus (CMV), Epstein-Barr virus (EBV)] or the identification of lymphocyte clonality. The latter application, however, does not detect specific DNA sequences, but rather Ig-κ and Ig-λ light-chain RNA.
(m)FISH Protocols
The following protocols are dedicated to (m)FISH, i.e., the use of fluorescence-labeled hybridization probes, with FFPE tissues (Basic Protocol 1) or cytological specimens (Basic Protocol 2). Oligomarker ISH using conventional (i.e., non-fluorescent) chromogen-tagged probes can also be used, although the visualization procedure differs from that using fluorochrome-labeled probes.
NOTE : All protocols involving animals must be reviewed and approved by the appropriate Animal Care and Use Committee and must follow regulations for the care and use of laboratory animals. Appropriate informed consent is necessary for obtaining and use of human study material.
Basic Protocol 1: (m)FISH APPLIED TO FORMALDEHYDE-FIXED PARAFFIN-EMBEDDED TISSUES
m(FISH) Procedure with FFPE Tissues
The FISH procedure basically comprises the following steps: slide preparation, specimen pretreatment, probe hybridization, and slide mounting. Afterward, the hybridization result is microscopically visualized. FFPE tissue sections can be prepared using a microtome. A thickness of ∼4 µm ensures the slicing of as few cell nuclei as possible. (Sliced nuclei are likely to show a loss of hybridization signals.) Tissue sections are immobilized preferably on charged glass microscopy slides that facilitate adherence. For pretreatment, tissue section slides are deparaffinized, rehydrated, and subsequently incubated in pretreatment solution (PS), which commonly contains HCl and NaSCN. This preparation is followed by enzymatic digestion with a pepsin solution. Via this pretreatment, disulfide bonds become broken, and DNA-bound proteins, i.e., histones, dissociate from the genomic DNA. Altogether, the preparation makes the DNA accessible to the hybridization probes and promotes FISH signal evaluation by reducing autofluorescence.
Alternative Protocols, Probes, and Probe Kits Designed to Accelerate the Preparation of FFPE FISH Specimens
Over the past few years, manufacturers have modified and optimized hybridization probes and probe kits (including a variety of preparation buffers and solutions) in order to shorten the turnaround preparation time for HER2 diagnostics from 1.5 days to ∼0.5 days. Two prominent, systematically validated, and commercially distributed product lines are the HER2 IQFISH pharmDx (Dako Agilent Technologies) and the FlexISH (ZytoVision) probe kits. Both kits enable the user to run the respective hybridization assay significantly faster than the traditional FISH assays. Qualitative and quantitative FISH results derived from “fast” and “traditional” preparations have been shown to excellently concord, which holds true for both the IQFISH (Franchet et al., 2014; Viale et al., 2016) and the FlexISH (Brockhoff et al., 2016) systems. An additional benefit of the IQFISH hybridization system is that it avoids the use of toxic formamide, which is commonly used for blocking against repetitive DNA sequences and to prevent nonspecific hybridization. By comparison, a significant advantage of the FlexISH product is the possibility of varying or adapting the preparation procedure and turnaround time based on the lab requirements. In other words, the FlexISH protocol allows application of both conventional and expedited preparation procedures, which might contribute to optimizing the flow of the daily workload in a diagnostic lab.
Basic Protocol 2: (m)FISH APPLIED TO CYTOLOGICAL SPECIMENS
Instead of using FFPE tissues (Basic Protocol 1), (m)FISH can, for diagnostic or research purposes, also be applied to cytological slide specimens derived from cell suspensions or bodily fluids (e.g., ascites, urine samples). For this purpose, a sufficient number of cells needs to be spun and immobilized on coated and positively charged slides. After fixation, cytospin slides can be stored at –20°C until further processing.
Additional Materials (also see Basic Protocol 1)
- Phosphate-buffered saline (PBS)
- Previously washed (with PBS) cell suspension
- 1× SSC buffer
- 1% and 4% (v/v) formalin solutions
- Ethanol series (prepared according to Basic Protocol 1, step 1)
-
- Cytospin funnels (Thermo Scientific)
- Positively charged microscope slides (e.g., Menzel Superfrost, Thermo Fisher Scientific, cat. no. MZ-00011)
First day
Cell immobilization
1.Prepare Cytospin funnels according to the manufacturer's manual, using positively charged microscope slides.
2.Preload assembled Cytospin funnels with 100 µl PBS.
3.Transfer 20,000 cells out of the previously washed (with PBS) cell suspension into the Cytospin funnels.
4.Spin cells onto the slides for 10 min at 600 × g.
5.Dismantle Cytospin funnels and release slides. Continue to processing immediately to avoid drying out the specimens.
6.Transfer slides to a 4% formalin solution in a staining jar or bath and incubate for 10 min.
7.Store specimens ≤1 year at −20°C or continue to further processing immediately.
Specimen pretreatment
8.Wash specimens twice in 1× SSC buffer for 2 min at 73°C.
9.Incubate in pepsin solution for 20 min at 37°C.
10.Incubate in 1× wb-3 (SSC buffer supplemented with 0.1% Igepal) for 5 min in glass cuvettes.
11.Incubate in 1% formalin solution for 5 min in glass cuvettes.
12.Incubate in 1× wb-3 for 5 min in glass cuvettes.
13.Wash in deionized water for 1 min in glass cuvettes.
14.Dehydrate using an ascending ethanol series (70%, 90%, and 100%) for 1 min each in glass cuvettes.
15.Let specimens air-dry.
Denaturation and hybridization
16.Cover the specimens (lying in horizontal position) with hybridization probe solution.
17.Cover specimens with coverslips and seal using rubber cement. Incubate at 75°C on a hot plate or hybridizer for 10 min.
18.Transfer to a hybridizer or humidity chamber at 37°C and incubate overnight.
Second day
Solution preparation
19.Pre-warm washing buffers (1× wb-1, wb-2, and wb-3) to 73°C in a water bath.
Post-hybridization processing
20.Incubate slides in warm 1× wb-1 for 2 min at room temperature in glass cuvettes.
21.Incubate slides in warm 2× wb-1 for 2 min at room temperature in glass cuvettes.
22.Incubate slides in warm 3× wb-1 for 2 min at room temperature in glass cuvettes.
23.Air-dry specimens while protecting them from light.
24.Cover specimens with DAPI-containing mounting solution.
25.Incubate the slides in the dark to enable the penetration of DAPI into cell nuclei.
Signal detection
26.After processing, examine the specimens via a fluorescence microscope with immersion oil. If not immediately used, store the specimens in the dark for short-term storage or at 2° to 8°C for long-term storage.
COMMENTARY
Background Information
Interphase FISH is a relatively easy and fast executable approach that in many cases complements (immuno-)histological diagnosis of already recognized or putative malignant tissues or cytologies. In addition to histological evaluation of the tissue architecture (usually done by a pathologist) and immunohistological assessment of cellular phenotypes (i.e., basically biomarker/antigene expression), it provides additional information about (potential) genomic alterations, among them gene amplifications, chromosomal translocations and rearrangements, or whole chromosomal losses or multiplications, the last commonly called polysomies. In particular, the fluorescence-based approach allows the quantitative assessment of multiple chromosomal markers at the same time and facilitates their evaluation on a cell-by-cell basis. Moreover, careful inspection of multi-marker FISH specimens enables the assessment of potential cell and tissue heterogeneity within tumor tissues. Even though FISH is a powerful complementary diagnostic tool that is very much suitable for the identification of known disease-specific chromosomal alterations in tumor tissues, it is not a substitute for modern, advanced molecular-pathological techniques, such as quantitative and multiplex PCRs, panel or next-generation sequencing, or multi-gene expression analyses. Likewise, single-nucleotide or oligonucleotide mutations cannot be identified by the hybridization of larger chromosomal regions. In contrast to those and other approaches, the information revealed by chromogenic and fluorescence-based ISH is derived from single cell inspections and is not accompanied by loss of tissue architecture and morphology.
A certain methodical advantage of FISH is that the identification of chromosomal rearrangements, as gains or losses do usually not require extra (positive or negative) controls because hybridization results are, in most cases, readily apparent, quantitative, and thus unequivocal. Nevertheless, the inspection of FISH specimens by two independent observers is generally recommended even though FISH images can now be evaluated (semi-)automatically. Furthermore, the inspection of sufficiently large and different tissue areas is essential to make sure that potential heterogeneity is not overlooked and ignored. Another advantage is that, once established, appropriate FISH preparation protocols can be applied to a variety of tissue types in the same way and can be practically transferred, one to one, to the use of different hybridization probes, irrespective of which chromosomal aberration (e.g., amplification, translocation) is the subject of interest. Last but not least, the financial outlay is quite reasonable.
Critical Parameters and Troubleshooting
Pitfalls, tips, and troubleshooting
Make sure to dry sections completely prior to probe application in Basic Protocols 1 and 2. Residual moisture may adversely affect signal intensity and/or tissue morphology.
When adding probe solution to each slide, make sure that the specimen gets completely covered. This prevents blank (i.e., not hybridized) tissue areas and enables the identification of potential cellular heterogeneity.
Remove probe-covered specimens from direct light and avoid long light exposure. This prevents fading or bleaching of the fluorochrome. This issue is not relevant when using conventional (non-fluorescent) chromogens.
Avoid the formation of air bubbles when covering slides with coverslips. Air bubbles cause uncovered tissue areas and irregular probe hybridization. Sealing of the coverslips by using rubber cement (or a similar product) prevents slipping of the coverslips out of position and, even more importantly, prevents evaporation of the probe solution.
Signal visualization, evaluation (counting), and data interpretation
Microscopy of (F)ISH specimens is the basis for the assessment of hybridization spots for Basic Protocols 1 and 2. In the case of FISH, make sure that the microscope is equipped with appropriate single- and dual-color filters. When analyzing tissues, make sure to evaluate a relevant tissue area of interest. Getting an initial overview by using a moderate magnification helps to identify the region of interest. When using a higher magnification (e.g., 40×, 63×), make sure to evaluate non-sliced nuclei and to assess all hybridization spots by carefully focusing across the 3D structure of cell nuclei.
Verify the quality of hybridization by using positive-control specimens whenever available.
Try to count every single hybridization spot in each cell nucleus. Count signal nucleus by nucleus, not marker by marker. When fluorochrome-labeled probes are applied, the use of dual- or tri-color fluorescence filters enormously facilitates nucleus-by-nucleus signal counting.
A number of hybridization spots can appear in clusters. Hybridization signals should be interpreted as individual spots if the distance between the spots amounts to about one signal diameter or more.
Do not evaluate overlapping, apparently sliced, or over-digested/denaturized nuclei.
For data interpretation (especially in diagnostics), apply official (i.e., binding) guidelines, if available.
Understanding Results
Interpretation of data revealed by (F)ISH cannot be generalized and depends on the probes applied, i.e., probes designed to detect gene copy numbers, translocations, rearrangements, and so forth. Hybridization spots and patterns can be easily and reliably rated when compared to those derived from control tissues or cells (e.g., normal, non-malignant samples). However, control specimens are not always required because modifications such as translocations or amplifications can also be identified without comparison to specimens known to not harbor any genomic alteration. A major advantage of a fluorescence-based approach over an chromogenic approach is that FISH results are inherently unequivocal, quantitative, and less user dependent. Nevertheless, the inspection of FISH specimens by two independent observers is strongly recommended.
Time Considerations
The typical interphase (m)FISH procedure performed on FFPE specimens (Basic Protocols 1 and 2) takes about two half days (including overnight incubation). Processing can be accelerated by the use of probes and protocols optimized for this purpose. Nevertheless, an established, more common approach can probably not be finished within <1 day.
Acknowledgments
The author thanks Britta Meyer and Sven Hauke (both ZytoVision GmbH, Bremerhaven, Germany) for their support.
Open access funding enabled and organized by Projekt DEAL.
Author Contributions
Gero Brockhoff : Conceptualization; data curation; investigation; methodology; project administration; validation; visualization; writing—original draft.
Conflict of Interest
The author declares no conflict of interest.
Open Research
Data Availability Statement
Data sharing is not applicable to this article as no new data were created or analyzed in this work.
Literature Cited
- Azofeifa, J., Fauth, C., Kraus, J., Maierhofer, C., Langer, S., Bolzer, A., Reichman, J., Schuffenhauer, S., & Speicher, M. R. (2000). An optimized probe set for the detection of small interchromosomal aberrations by use of 24-color FISH. American Journal of Human Genetics , 66(5), 1684–1688. https://doi.org/10.1086/302875
- Baerlocher, G. M., Vulto, I., de Jong, G., & Lansdorp, P. M. (2006). Flow cytometry and FISH to measure the average length of telomeres (flow FISH). Nature Protocols , 1(5), 2365–2376. https://doi.org/10.1038/nprot.2006.263
- Bayani, J., & Squire, J. A. (2004a). Fluorescence in situ hybridization (FISH). Current Protocols in Cell Biology , 23(1), 22.4.1–22.4.52. https://doi.org/10.1002/0471143030.cb2204s23
- Bayani, J., & Squire, J. A. (2004b). Traditional banding of chromosomes for cytogenetic analysis. Current Protocols in Cell Biology , 23(1), 22.3.1–22.3.7. https://doi.org/10.1002/0471143030.cb2203s23
- Bayani, J., & Squire, J. A. (2005). Comparative genomic hybridization. Current Protocols in Cell Biology , 25(1), 22.6.1–22.6.15. https://doi.org/10.1002/0471143030.cb2206s25
- Bilous, M., Morey, A., Armes, J., Cummings, M., & Francis, G. (2006). Chromogenic in situ hybridisation testing for HER2 gene amplification in breast cancer produces highly reproducible results concordant with fluorescence in situ hybridisation and immunohistochemistry. Pathology , 38(2), 120–124. https://doi.org/10.1080/00313020600561518
- Bjerregaard, V. A., Özer, Ö., Hickson, I. D., & Liu, Y. (2018). The detection and analysis of chromosome fragile sites. Methods in Molecular Biology , 1672, 471–482. https://doi.org/10.1007/978-1-4939-7306-4_31
- Brockhoff, G., Bock, M., Zeman, F., & Hauke, S. (2016). The FlexISH assay brings flexibility to cytogenetic HER2 testing. Histopathology , 69(4), 635–646. https://doi.org/10.1111/his.12974
- Daniely, M., Rona, R., Kaplan, T., Olsfanger, S., Elboim, L., Freiberger, A., Lew, S., & Leibovitch, I. (2007). Combined morphologic and fluorescence in situ hybridization analysis of voided urine samples for the detection and follow-up of bladder cancer in patients with benign urine cytology. Cancer , 111(6), 517–524. https://doi.org/10.1002/cncr.23119
- Dawood, S., Broglio, K., Buzdar, A. U., Hortobagyi, G. N., & Giordano, S. H. (2010). Prognosis of women with metastatic breast cancer by HER2 status and trastuzumab treatment. An institutional-based review. Journal of Clinical Oncology , 28(1), 92–98. https://doi.org/10.1200/JCO.2008.19.9844
- Denzinger, S., Stoehr, R., Schwarz, S., Eichenseher, N., Brockhoff, G., Obermann, E. C., Knuechel, R., Blaszyk, H., Hartmann, A., & Wild, P. J. (2007). Low level STK15 amplification in histologically benign urothelium of patients with bladder cancer adversely predicts patient outcome following cystectomy. International Journal of Oncology , 31(4), 793–802. https://doi.org/10.3892/ijo.31.4.793
- Donaldson, A. R., Shetty, S., Wang, Z., Rivera, C. L., Portier, B. P., Budd, G. T., Downs-Kelly, E., Lanigan, C. P., & Calhoun, B. C. (2017). Impact of an alternative chromosome 17 probe and the 2013 American Society of Clinical Oncology and College of American Pathologists guidelines on fluorescence in situ hybridization for the determination of HER2 gene amplification in breast cancer. Cancer , 123(12), 2230–2239. https://doi.org/10.1002/cncr.30592
- Franchet, C., Filleron, T., Cayre, A., Mounié, E., Penault-Llorca, F., Jacquemier, J., Macgrogan, G., Arnould, L., & Lacroix-Triki, M. (2014). Instant-quality fluorescence in-situ hybridization as a new tool for HER2 testing in breast cancer. A comparative study. Histopathology , 64(2), 274–283. https://doi.org/10.1111/his.12247
- Frithiof, H., Aaltonen, K., & Ryden, L. (2016). A FISH-based method for assessment of HER-2 amplification status in breast cancer circulating tumor cells following CellSearch isolation. OncoTargets and Therapy , 9, 7095–7103. https://doi.org/10.2147/OTT.S118502
- Green, I. F., & Zynger, D. L. (2015). Institutional quality assurance for breast cancer HER2 immunohistochemical testing. Identification of outlier results and impact of simultaneous fluorescence in situ hybridization cotesting. Human Pathology , 46(12), 1842–1849. https://doi.org/10.1016/j.humpath.2015.08.001
- Hanna, W. M., Rüschoff, J., Bilous, M., Coudry, R. A., Dowsett, M., Osamura, R. Y., Penault-Llorca, F., van de Vijver, M., & Viale, G. (2014). HER2 in situ hybridization in breast cancer. Clinical implications of polysomy 17 and genetic heterogeneity. Modern Pathology , 27(1), 4–18. https://doi.org/10.1038/modpathol.2013.103
- Hauser-Kronberger, C., & Dandachi, N. (2004). Comparison of chromogenic in situ hybridization with other methodologies for HER2 status assessment in breast cancer. Journal of Molecular Histology , 35(6), 647–653. https://doi.org/10.1007/s10735-004-2186-6
- He, C. (2013). Correlation of human epidermal growth factor receptor 2 expression with clinicopathological characteristics and prognosis in gastric cancer. World Journal of Gastroenterology , 19(14), 2171–2178. https://doi.org/10.3748/wjg.v19.i14.2171
- Hoffmann, M., Pasch, S., Schamberger, T., Maneck, M., Möhlendick, B., Schumacher, S., Brockhoff, G., Knoefel, W. T., Izbicki, J., Polzer, B., Stoecklein, N. H., & Klein, C. A. (2018). Diagnostic pathology of early systemic cancer. ERBB2 gene amplification in single disseminated cancer cells determines patient survival in operable esophageal cancer. International Journal of Cancer , 142(4), 833–843. https://doi.org/10.1002/ijc.31108
- Holzschuh, M.-A., Czyz, Z., Hauke, S., Inwald, E. C., Polzer, B., & Brockhoff, G. (2017). HER2 FISH results in breast cancers with increased CEN17 signals using alternative chromosome 17 probes - reclassifying cases in the equivocal category. Histopathology , 71, 610–625. https://doi.org/10.1111/his.13253
- Ignatiadis, M., Rothé, F., Chaboteaux, C., Durbecq, V., Rouas, G., Criscitiello, C., Metallo, J., Kheddoumi, N., Singhal, S. K., Michiels, S., Veys, I., Rossari, J., Larsimont, D., Carly, B., Pestrin, M., Bessi, S., Buxant, F., Liebens, F., Piccart, M., & Sotiriou, C. (2011). HER2-positive circulating tumor cells in breast cancer. PLOS ONE , 6(1), e15624. https://doi.org/10.1371/journal.pone.0015624
- Kato, N., Itoh, H., Serizawa, A., Hatanaka, Y., Umemura, S., & Osamura, R. Y (2010). Evaluation of HER2 gene amplification in invasive breast cancer using a dual-color chromogenic in situ hybridization (dual CISH). Pathology International , 60(7), 510–515. https://doi.org/10.1111/j.1440-1827.2010.02553.x
- Knudson, R. A., Shearer, B. M., & Ketterling, R. P. (2007). Automated Duet spot counting system and manual technologist scoring using dual-fusion fluorescence in situ hybridization (D-FISH) strategy: Comparison and application to FISH minimal residual disease testing in patients with chronic myeloid leukemia. Cancer Genetics and Cytogenetics , 175(1), 8–18. https://doi.org/10.1016/j.cancergencyto.2006.12.006
- Koudelakova, V., Berkovcova, J., Trojanec, R., Vrbkova, J., Radova, L., Ehrmann, J., Kolar, Z., Melichar, B., & Hajduch, M. (2015). Evaluation of HER2 gene status in breast cancer samples with indeterminate fluorescence in situ hybridization by quantitative real-time PCR. The Journal of Molecular Diagnostics , 17(4), 446–455. https://doi.org/10.1016/j.jmoldx.2015.03.007
- Koudelakova, V., Trojanec, R., Vrbkova, J., Donevska, S., Bouchalova, K., Kolar, Z., Varanasi, L., & Hajduch, M. (2016). Frequency of chromosome 17 polysomy in relation to CEP17 copy number in a large breast cancer cohort. Genes, Chromosomes & Cancer, 55(5), 409–417. https://doi.org/10.1002/gcc.22337
- Krishnamurthy, S., Bischoff, F., Ann Mayer, J., Wong, K., Pham, T., Kuerer, H., Lodhi, A., Bhattacharyya, A., Hall, C., & Lucci, A. (2013). Discordance in HER2 gene amplification in circulating and disseminated tumor cells in patients with operable breast cancer. Cancer Medicine , 2(2), 226–233. https://doi.org/10.1002/cam4.70
- Langer, S., Kraus, J., Jentsch, I., & Speicher, M. R. (2004). Multicolor chromosome painting in diagnostic and research applications. Chromosome Research , 12(1), 15–23. https://doi.org/10.1023/B:CHRO.0000009326.21752.88
- Lee, C., Lemyre, E., Miron, P. M., & Morton, C. C. (2001). Multicolor fluorescence in situ hybridization in clinical cytogenetic diagnostics. Current Opinion in Pediatrics , 13(6), 550–555. https://doi.org/10.1097/00008480-200112000-00010
- Liehr, T., Othman, M. A. K., & Rittscher, K. (2017). Multicolor karyotyping and fluorescence in situ hybridization-banding (MCB/mBAND). Methods in Molecular Biology , 1541, 181–187. https://doi.org/10.1007/978-1-4939-6703-2_16
- Liehr, T., Starke, H., Heller, A., Kosyakova, N., Mrasek, K., Gross, M., Karst, C., Steinhaeuser, U., Hunstig, F., Fickelscher, I., Kuechler, A., Trifonov, V., Romanenko, S. A., & Weise, A. (2006). Multicolor fluorescence in situ hybridization (FISH) applied to FISH-banding. Cytogenetic and Genome Research , 114(3–4), 240–244. https://doi.org/10.1159/000094207
- Li-Ning-T, E., Ronchetti, R., Torres-Cabala, C., & Merino, M. J. (2005). Role of chromogenic in situ hybridization (CISH) in the evaluation of HER2 status in breast carcinoma. Comparison with immunohistochemistry and FISH. International Journal of Surgical Pathology , 13(4), 343–351. https://doi.org/10.1177/106689690501300406
- Loring, P., Cummins, R., O'Grady, A., & Kay, E. W. (2005). HER2 positivity in breast carcinoma. A comparison of chromogenic in situ hybridization with fluorescence in situ hybridization in tissue microarrays, with targeted evaluation of intratumoral heterogeneity by in situ hybridization. Applied Immunohistochemistry & Molecular Morphology, 13(2), 194–200. https://doi.org/10.1097/01.pai.0000132189.01233.6d
- Lottner, C., Schwarz, S., Diermeier, S., Hartmann, A., Knuechel, R., Hofstaedter, F., & Brockhoff, G. (2005). Simultaneous detection of HER2/neu gene amplification and protein overexpression in paraffin-embedded breast cancer. The Journal of Pathology , 205(5), 577–584. https://doi.org/10.1002/path.1742
- MacLeod, R. A. F., Kaufmann, M. E., & Drexler, H. G. (2017). Cytogenetic harvesting of cancer cells and cell lines. Methods in Molecular Biology , 1541, 43–58. https://doi.org/10.1007/978-1-4939-6703-2_5
- Maguire, O., Wallace, P. K., & Minderman, H. (2016). Fluorescent in situ hybridization in suspension by imaging flow cytometry. Methods in Molecular Biology , 1389, 111–126. https://doi.org/10.1007/978-1-4939-3302-0_7
- Maiti, S. N. (2016). Measurement of average telomere length in ex vivo expanded natural killer cells by fluorescence in situ hybridization (FISH) and flow cytometry. Methods in Molecular Biology , 1441, 57–63. https://doi.org/10.1007/978-1-4939-3684-7_5
- Manaresi, E., Bua, G., Bonvicini, F., & Gallinella, G. (2015). A flow-FISH assay for the quantitative analysis of parvovirus B19 infected cells. Journal of Virological Methods , 223, 50–54. https://doi.org/10.1016/j.jviromet.2015.07.013
- Marchiò, C., Lambros, M. B., Gugliotta, P., Di Cantogno, L. V., Botta, C., Pasini, B., Tan, D. S., Mackay, A., Fenwick, K., Tamber, N., Bussolati, G., Ashworth, A., Reis-Filho, J. S., & Sapino, A. (2009). Does chromosome 17 centromere copy number predict polysomy in breast cancer? A fluorescence in situ hybridization and microarray-based CGH analysis. The Journal of Pathology , 219(1), 16–24. https://doi.org/10.1002/path.2574
- Martinez, G., Gillois, P., le Mitouard, M., Borye, R., Esquerré-Lamare, C., Satre, V., Bujan, L., & Hennebicq, S. (2013). FISH and tips: A large scale analysis of automated versus manual scoring for sperm aneuploidy detection. Basic and Clinical Andrology , 23, 13. https://doi.org/10.1186/2051-4190-23-13
- Mccormick, S. R., Lillemoe, T. J., Beneke, J., Schrauth, J., & Reinartz, J. (2002). HER2 assessment by immunohistochemical analysis and fluorescence in situ hybridization. Comparison of HercepTest and PathVysion commercial assays. American Journal of Clinical Pathology , 117(6), 935–943. https://doi.org/10.1309/3643-F955-7Q6B-EWWL
- Minderman, H., Humphrey, K., Arcadi, J. K., Wierzbicki, A., Maguire, O., Wang, E. S., Block, A. W., Sait, S. N. J., George, T. C., & Wallace, P. K. (2012). Image cytometry based detection of aneuploidy by fluorescence in situ hybridization in suspension. Cytometry Part A , 81A(9), 776–784. https://doi.org/10.1002/cyto.a.22101
- Miyake, M., Hori, S., Fujii, T., Nishimura, N., Oda, Y., Miyamoto, T., Tomizawa, M., Shimizu, T., Ohnishi, K., Morizawa, Y., Gotoh, D., Nakai, Y., Torimoto, K., Tanaka, N., & Fujimoto, K. (2023). Clinical practice of UroVysion® urine test in patients with bladder carcinoma in situ treated with intravesical Bacillus Calmette-Guerin. Japanese Journal of Clinical Oncology , 53, 629–632. https://doi.org/10.1093/jjco/hyad029
- Nakamura, Y., Itoh, Y., Kakegawa, E., Uchida, Y., Ichimura, T., & Sasaki, A. (2022). Biallelic BCL6 rearrangements by dual t(3;14)(q27;q32) and t(3;22)(q27;q11) translocations in diffuse large B-cell lymphoma. Journal of Clinical and Experimental Hematopathology , 62(4), 268–272. https://doi.org/10.3960/jslrt.22031
- Narath, R., Lörch, T., Rudas, M., & Ambros, P. F. (2004). Automatic quantification of gene amplification in clinical samples by IQ-FISH. Cytometry Part B , 57B(1), 15–22. https://doi.org/10.1002/cyto.b.10058
- Nishimura, R., Kagawa, A., Tamogami, S., Kojima, K., Satou, M., Yamashita, N., Teramoto, N., & Aogi, K. (2016). Correlation of HER2 gene status assessment by fluorescence in situ hybridization between histological sections and cytological specimens of breast cancer. Breast Cancer , 23(2), 211–215. https://doi.org/10.1007/s12282-014-0552-0
- Ohno, H., Maekawa, F., Nakagawa, M., Chagi, Y., Nakagawa, M., Kishimori, C., Fukutsuka, K., Hayashida, M., Takeoka, K., Maruyama, W., Ukyo, N., & Sumiyoshi, S. (2022). Two cases of primary diffuse large B-cell lymphoma of the CNS associated with t(8;14)(q24;q32) or t(3;14)(q27;q32) identified by G-banding and fluorescence in situ hybridization applied to metaphase spreads. Journal of Clinical and Experimental Hematopathology , 62(4), 242–248. https://doi.org/10.3960/jslrt.22019
- Pazhoomand, R., Keyhan, E., Banan, M., Najmabad, H., Karimlou, M., Khodadad, F., Iraniparast, A., Feiz, F., Majidzadeh, K., Bahman, I., Moghadam, F. A., Sobhani, A. M., Abedin, S. S., Muhammadnejad, A., & Behjat, F. (2013). Detection of HER2 status in breast cancer: Comparison of current methods with MLPA and real-time RT-PCR. Asian Pacific Journal of Cancer Prevention , 14(12), 7621–7628. https://doi.org/10.7314/APJCP.2013.14.12.7621
- Perez, E. A., Reinholz, M. M., Hillman, D. W., Tenner, K. S., Schroeder, M. J., Davidson, N. E., Martino, S., Sledge, G. W., Harris, L. N., Gralow, J. R., Dueck, A. C., Ketterling, R. P., Ingle, J. N., Lingle, W. L., Kaufman, P. A., Visscher, D. W., & Jenkins, R. B. (2010). HER2 and chromosome 17 effect on patient outcome in the N9831 adjuvant trastuzumab trial. Journal of Clinical Oncology , 28(28), 4307–4315. https://doi.org/10.1200/JCO.2009.26.2154
- Pergament, E., Xien Chen, P., Thangavelu, M., & Fiddler, M. (2000). The clinical application of interphase FISH in prenatal diagnosis. Prenatal Diagnosis , 20(3), 215–220. https://doi.org/10.1002/(SICI)1097-0223(200003)20:3%3c215::AID-PD785%3e3.0.CO;2-X
- Pothos, A., Plastira, K., Plastiras, A., Vlachodimitropoulos, D., Goutas, N., & Angelopoulou, R. (2008). Comparison of chromogenic in situ hybridisation with fluorescence in situ hybridisation and immunohistochemistry for the assessment of her-2/neu oncogene in archival material of breast carcinoma. Acta Histochemica et Cytochemica , 41(3), 59–64. https://doi.org/10.1267/ahc.07029
- Ranjbaran, R., Okhovat, M. A., Abbasi, M., Moezzi, L., Aboualizadeh, F., Amidzadeh, Z., Golafshan, H. A., Behzad-Behbahani, A., & Sharifzadeh, S. (2016). Detection of t(9;22) b2a2 fusion transcript by flow cytometry. International Journal of Laboratory Hematology , 38(4), 403–411. https://doi.org/10.1111/ijlh.12515
- Salam, D. S. D. A., Thit, E. E., Teoh, S. H., Tan, S. Y., Peh, S. C., & Cheah, S.-C. (2020). C-MYC, BCL2 and BCL6 translocation in B-cell non-Hodgkin lymphoma cases. Journal of Cancer , 11(1), 190–198. https://doi.org/10.7150/jca.36954
- Sassen, A., Rochon, J., Wild, P., Hartmann, A., Hofstaedter, F., Schwarz, S., & Brockhoff, G. (2008). Cytogenetic analysis of HER1/EGFR, HER2, HER3 and HER4 in 278 breast cancer patients. Breast Cancer Research , 10(1), R2. https://doi.org/10.1186/bcr1843
- Schwarz, S., Rechenmacher, M., Filbeck, T., Knuechel, R., Blaszyk, H., Hartmann, A., & Brockhoff, G. (2008). Value of multicolour fluorescence in situ hybridisation (UroVysion) in the differential diagnosis of flat urothelial lesions. Journal of Clinical Pathology , 61(3), 272–277. https://doi.org/10.1136/jcp.2007.049684
- Schwarz, S., Rechenmacher, M., Lottner, C., Brockhoff, G., Hartmann, A., Langer, S., & Knüchel, R. (2004). FISH-untersuchungen an multi-tissue-arrays zur differenzierung flacher läsionen des urothels. Verhandlungen der Deutschen Gesellschaft fur Pathologie , 88, 184–193.
- Shao, L., Miller, S., Keller-Ramey, J., Zhang, Y., & Roulston, D. (2015). Cytogenetic, fluorescence in situ hybridization, and genomic array characterization of chronic myeloid leukemia with cryptic BCR-ABL1 fusions. Cancer Genetics , 208(7–8), 396–403. https://doi.org/10.1016/j.cancergen.2015.04.006
- Slamon, D. J., Clark, G. M., Wong, S. G., Levin, W. J., Ullrich, A., & Mcguire, W. L. (1987). Human breast cancer: Correlation of relapse and survival with amplification of the HER-2/neu oncogene. Science , 235(4785), 177–182. https://doi.org/10.1126/science.3798106
- Slamon, D. J., Leyland-Jones, B., Shak, S., Fuchs, H., Paton, V., Bajamonde, A., Fleming, T., Eiermann, W., Wolter, J., Pegram, M., Baselga, J., & Norton, L. (2001). Use of chemotherapy plus a monoclonal antibody against HER2 for metastatic breast cancer that overexpresses HER2. The New England Journal of Medicine , 344(11), 783–792. https://doi.org/10.1056/NEJM200103153441101
- Smith, G. D., Riding, M., Oswald, K., & Bentz, J. S. (2010). Integrating a FISH imaging system into the cytology laboratory. CytoJournal , 7, 3. https://doi.org/10.4103/1742-6413.62258
- Swennenhuis, J. F., Tibbe, A. G. J., Levink, R., Sipkema, R. C. J., & Terstappen, L. W. M. M. (2009). Characterization of circulating tumor cells by fluorescence in situ hybridization. Cytometry Part A , 75A(6), 520–527. https://doi.org/10.1002/cyto.a.20718
- Tepperberg, J., Pettenati, M. J., Rao, P. N., Lese, C. M., Rita, D., Wyandt, H., Gersen, S., White, B., & Schoonmaker, M. M. (2001). Prenatal diagnosis using interphase fluorescence in situ hybridization (FISH): 2-year multi-center retrospective study and review of the literature. Prenatal Diagnosis , 21(4), 293–301. https://doi.org/10.1002/pd.57
- Viale, G., Paterson, J., Bloch, M., Csathy, G., Allen, D., Dell'orto, P., Kjærsgaard, G., Levy, Y. Y., & Jørgensen, J. T. (2016). Assessment of HER2 amplification status in breast cancer using a new automated HER2 IQFISH pharmDx (Dako Omnis) assay. Pathology, Research and Practice , 212(8), 735–742. https://doi.org/10.1016/j.prp.2016.06.002
- Virgili, A., & Nacheva, E. P. (2010). Genomic amplification of BCR/ABL1 and a region downstream of ABL1 in chronic myeloid leukaemia. A FISH mapping study of CML patients and cell lines. Molecular Cytogenetics , 3, 15. https://doi.org/10.1186/1755-8166-3-15
- Wolff, A. C., Hammond, M. E. H., Allison, K. H., Harvey, B. E., Mcshane, L. M., & Dowsett, M. (2018). HER2 testing in breast cancer: American Society of Clinical Oncology/College of American Pathologists clinical practice guideline focused update summary. Journal of Oncology Practice , 14(7), 437–441. https://doi.org/10.1200/JOP.18.00206
- Wolff, A. C., Hammond, M. E. H., Hicks, D. G., Dowsett, M., Mcshane, L. M., Allison, K. H., Allred, D. C., Bartlett, J. M. S., Bilous, M., Fitzgibbons, P., Hanna, W., Jenkins, R. B., Mangu, P. B., Paik, S., Perez, E. A., Press, M. F., Spears, P. A., Vance, G. H., Viale, G., & Hayes, D. F. (2013). Recommendations for human epidermal growth factor receptor 2 testing in breast cancer: American Society of Clinical Oncology/College of American Pathologists clinical practice guideline update. Journal of Clinical Oncology , 31(31), 3997–4013. https://doi.org/10.1200/JCO.2013.50.9984
- Wolff, A. C., Hammond, M. E. H., Schwartz, J. N., Hagerty, K. L., Allred, D. C., Cote, R. J., Dowsett, M., Fitzgibbons, P. L., Hanna, W. M., Langer, A., Mcshane, L. M., Paik, S., Pegram, M. D., Perez, E. A., Press, M. F., Rhodes, A., Sturgeon, C., Taube, S. E., Tubbs, R., … Hayes, D. F. (2007). American Society of Clinical Oncology/College of American Pathologists guideline recommendations for human epidermal growth factor receptor 2 testing in breast cancer. Journal of Clinical Oncology , 25(1), 118–145. https://doi.org/10.1200/JCO.2006.09.2775
- Zhang, Y., Perez, T., Blondin, B., Du, J., Liu, P., Escarzaga, D., Coon, J. S., Morrison, L. E., & Pestova, K. (2014). Identification of FISH biomarkers to detect chromosome abnormalities associated with prostate adenocarcinoma in tumour and field effect environment. BMC Cancer , 14, 129. https://doi.org/10.1186/1471-2407-14-129
- Zhou, H., Yan, Y., & Song, L. (2016). The clinical application of fluorescence in situ hybridization in diagnosing urothelial carcinoma. Clinical Laboratory , 62(10), 2001–2009. https://doi.org/10.7754/Clin.Lab.2016.160323
- Zwaenepoel, K., Merkle, D., Cabillic, F., Berg, E., Belaud-Rotureau, M.-A., Grazioli, V., Herelle, O., Hummel, M., le Calve, M., Lenze, D., Mende, S., Pauwels, P., Quilichini, B., & Repetti, E. (2015). Automation of ALK gene rearrangement testing with fluorescence in situ hybridization (FISH): A feasibility study. Experimental and Molecular Pathology , 98(1), 113–118. https://doi.org/10.1016/j.yexmp.2015.01.005
Corrections
In this publication, the following corrections have been made:
In the figure legend 1, “cen11” and “green-labeled” have been replaced with the correct words.
The current version online now includes these corrections and may be considered the authoritative version of record.

