Chloroform-methanol protein precipitation from microalgae and Pierce BCA assay
Ying-Yu Hu, Christopher Lord, Zoe V Finkel
Abstract
Chlorophyll, phospholipids, sucrose, glycerol and some detergent in crude protein extracted from microalgae can interfere the Pierce BCA protein assay. In order to remove these interference, bead miller extracted protein is precipitated by chloroform-methanol prior to BCA assay. The resulting precipitation is dissolved into Sarcosine-Tris solution. Low limit of detection is about 5 ug/mL.
Steps
Reagent preparation
Tris buffer 5 (pH 8.0)
Add 500µL 1``8.0 Tris into 100 mL MilliQ water
20% Sarcosine
Dilute 2 part 30% N-lauroylsarcosine sodium salt with 1 part 5 (pH 8.0) Tris buffer
Protein precipitation
Thaw protein extract
Turn on refrigerate centrifuge
Equipment
| Value | Label |
|---|---|
| CENTRIFUGE 5430 R | NAME |
| Eppendorf | BRAND |
| MP2231000510 | SKU |
Turn on incubator/shaker, preheat to 37°C
Equipment
| Value | Label |
|---|---|
| SHAKING INCUBATOR | NAME |
| 71L | TYPE |
| Corning® LSE™ | BRAND |
| 6753 | SKU |
Prepare ice-bath
Well mix the extract and then transfer 100µL of extract to 2 mL microtube (Abdos tubes give better precipitation results), in replicate.
Equipment
| Value | Label |
|---|---|
| Micro Centrifuge Tubes | NAME |
| Abdos | BRAND |
| P10203 | SKU |
In the fume hood, add 400µL methanol
Gently vortex for 0h 0m 30s by using a tube insert
Equipment
| Value | Label |
|---|---|
| VWR ANALOG VORTEX MIXER | NAME |
| VWR | BRAND |
| 10153-838 | SKU |
| With tube insert | SPECIFICATIONS |
In the fume hood, add 100µL chloroform
Gently vortex for 0h 0m 30s by using a tube insert
In the fume hood, add 300µL MilliQ
Gently vortex for 0h 0m 30s by using a tube insert
Incubate On ice for 0h 30m 0s
20000rcf,4°C
In the fume hood, remove upper phase by leaving about 250µL liquid
In the fume hood, add 300µL methanol
Gently mix the liquid until bottom layer disappear and the solution is homogenous.
20000rcf,4°C
In the fume hood, remove all solvent.
If pellet tends to be aspired with solvent, add another 300µL methanol, gently vortex, and 20000rcf,4°C
In the fume hood, remove most solvent by using 1000 uL pipet tip, and then remove the rest by using 100 uL pipet tip. Do not remove pellet with solvent.
Dry pellet in vacuum desiccator for at least 0h 30m 0s at Room temperature
BCA assay
Add 5µL 20% sarcosine and 95µL 5 (pH 8.0) Tris buffer to dry protein pellet, incubate at 37°C for 15 to 30 min.
Use tube insert, vortex all tubes for 15 to 30 min until pellet is completely re-dissolved.
BSA standard solutions
| A | B | C | D | E |
|---|---|---|---|---|
| SD1 | 5 | 95 | 0 | 0 |
| SD2 | 25 | 470 | 5 | 0.02 |
| SD3 | 25 | 463 | 12 | 0.048 |
| SD4 | 25 | 450 | 25 | 0.1 |
| SD5 | 25 | 425 | 50 | 0.2 |
| SD6 | 25 | 375 | 100 | 0.4 |
| SD7 | 25 | 275 | 200 | 0.8 |
| SD8 | 25 | 225 | 250 | 1 |
Vortex and then use reverse pipetting: transfer 100µL standard solutions into the corresponding tubes, except for SD1 (it has already been 100 uL).
Use the following formula to determine the total volume of working reagent (WR) required. Consider leaving several mL of extra volume:
(# standards + # samples) X (800µL) = total volume WR required
Prepare WR by mixing 50 parts of BCA reagent A with 1 part of BCA Reagent B in a 50 mL falcon tube
Use one tip and reverse pipetting: Add 800µL WR into each tube, make sure that the tip doesn't have contact with the solution, so that samples are not cross-contaminated.
Vortex each tube, shake and incubate at 37°C for 0h 30m 0s
Remove samples from the incubator.
Load samples into microplate in duplicate:
Equipment
| Value | Label |
|---|---|
| 96-Well Microplates | NAME |
| Polystyrene, Clear, | TYPE |
| Greiner Bio-One | BRAND |
| 82050-760 | SKU |
Shake for 5 s at 600 rpm in a continuous and high force modeRead endpoint 562 nm with a measurement time 100 ms Equipment
| Value | Label |
|---|---|
| Varioskan LUX Multimode Microplate Reader | NAME |
| Thermo Fisher | BRAND |
| VL0L00D0 | SKU |
Calculation
Subtract the average 562 nm absorbance measurement of the blank standard replicates from the 562 nm measurements of all other individual standard .
Subtract the average 562 nm absorbance measurement of the blank sample (filter) replicates from the 562 nm measurements of all other individual sample .
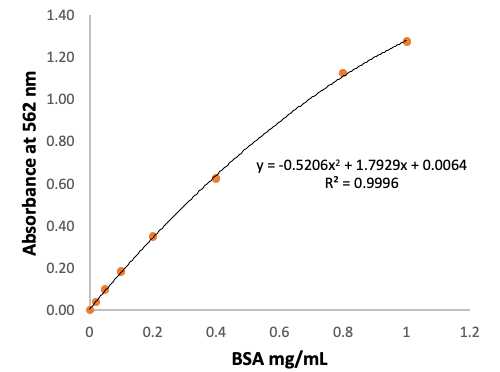
For the calculation convenience, plot BSA concentration (Conc) versus Corrected absorbance (Abs) to obtain a standard curve as following:
Conc_mg/mL = a X Abs^2 + b X Abs + c
Use the corrected measured absorbance of samples (Abs) to calculate the total protein concentration (Conc_mg/mL) from each sample.
Protein_mg/filter = Conc_mg/mL X PEB_mL
Where PEB is the volume of protein extraction buffer used to extract protein from microalgae sample.