ChIP-seq Library preparation
Vasso Makrantoni, Daniel Robertson, Adele L. Marston
Chromatin immunoprecipitation
Saccharomyces cerevisiae
Schizosaccharomyces pombe
Cohesin
Condensin
Mitosis
Meiosis
Scc1
Rec8
Brn1
Abstract
A plethora of biological processes like gene transcription, DNA replication, DNA recombination, and chromosome segregation are mediated through protein–DNA interactions. A powerful method for investigating proteins within a native chromatin environment in the cell is chromatin immunoprecipitation (ChIP). Combined with the recent technological advancement in next generation sequencing, the ChIP assay can map the exact binding sites of a protein of interest across the entire genome. Here we describe a-step-by step protocol for ChIP followed by library preparation for ChIP-seq from yeast cells.
Before start
Steps
ChIP-seq Library preparation
DNA End-Repair
Perform the blunting reaction using the following recipe: if making multiple libraries prepare a master mix of buffer, dNTPs, and enzyme, and then aliquot to the required ChIP purified DNA. The final volume should be 50 μl.
(a) 1ng–20ng (ideally 2 ng)
(b) 5µL
(c) 5µL
(d) 1µL
Incubate at 65Room temperature for 0h 45m 0s.
Perform a 1.6:1 AMPure XP selection by adding 80µL to the 50µL blunting reaction (see protocol "AMPure Purification Protocol").
Elute in 30µL and take 27.7µL to a new DNA LoBind Eppendorf.
“A”-Tailing Reaction
Use end-repaired DNA (from step 5) to perform “A”-tailing reaction using the following recipe: if making multiple libraries prepare a master mix of buffer, dATP, and enzyme, and then aliquot to the required end-repaired DNA. The final volume should be 30 μl.
(a) 27.7µL
(b) 3.3µL
(c) 1µL
(d) 1µL
Incubate at 37°C for 0h 30m 0s.
Heat-inactivate Klenow enzyme at 75°C for 0h 5m 0s.
Place reaction On ice for 0h 5m 0s.
Proceed immediately to adapter ligation reaction (next step).
Adapter Ligation Reaction
Use dA-tailed DNA (from previous step) to perform the adapter ligation reaction using the following recipe:
the final volume should be 70 μl. Use different barcoded adapters for each Input and IP sample.
(a) 33µL
(b) 35µL
(c) 1µL [15]
(d) 1µL
Incubate at Room temperature for 0h 25m 0s.
Perform a 1:1 AMPure selection by adding 70µL to the 70µL (see protocol "AMPure Purification Protocol").
Elute in 52µL. Transfer 50µL to a new DNA LoBind Eppendorf.
Perform another 1:1 AMPure selection by adding 50µL to the 50µL from the previous step.
Elute in 33µL and take up 30µL to a new DNA LoBind Eppendorf.
PCR Amplification Reaction
Transfer 10µL per sample (previous step) to a 0.2 ml PCR tube and set up the following PCR reaction On ice in a final volume of 50 μl.
(a) 10µL
(b) 10µL
(c) 4µL
(d) 2µL
(e) 1.5µL
(f) 0.5µL
(g) 22µL
Amplify DNA with the following PCR program:
| A | B | C |
|---|---|---|
| Temperature | Time | Cycles |
| 98 °C | 30 sec | |
| 98 °C | 10 sec | 12 -18 cycles |
| 65 °C | 30 sec | |
| 72 °C | 30 sec | |
| 72 °C | 5 min |
Double-Sided AMPure Selection and Library Elution
Perform a 0.65×:1 AMPure selection. To the 50µL (previous step) add 31.85µL, resuspend by pipetting and leave at Room temperature for 0h 10m 0s to bind.
Place 0.2 μl PCR tubes in a DynaMag-PCR magnet for 0h 5m 0s.
KEEP THE SUPERNATANT , this will contain fragments <300 bp. To 80µL add 50µL (adjust bead volume for smaller supernatant volumes) and perform AMPure selection according to steps in protocol "AMPure Purification Protocol".
Elute in 52µL and pipet 50µL to a new DNA LoBind Eppendorf.
Perform another 1:1 AMPure selection by adding 50µL to the 50µL from the previous step.
Elute in 33µL and transfer 30µL to a new LoBind Eppendorf.
Determine DNA library concentration by Qubit HS kit (use 2µL).
Run the library on a Bioanalyzer to determine average fragment size and general purity. Use a High Sensitivity DNA Kit (Agilent Technologies) as per manufacturer’s instructions. Fragments of the sequencing library should have a size range of 150–300 bp (Fig.2b, upper panel). If not pure, that is, adapter dimers are visible (Fig.2b, lower panel), perform 1:1 AMPure purification to remove small adapter dimers.
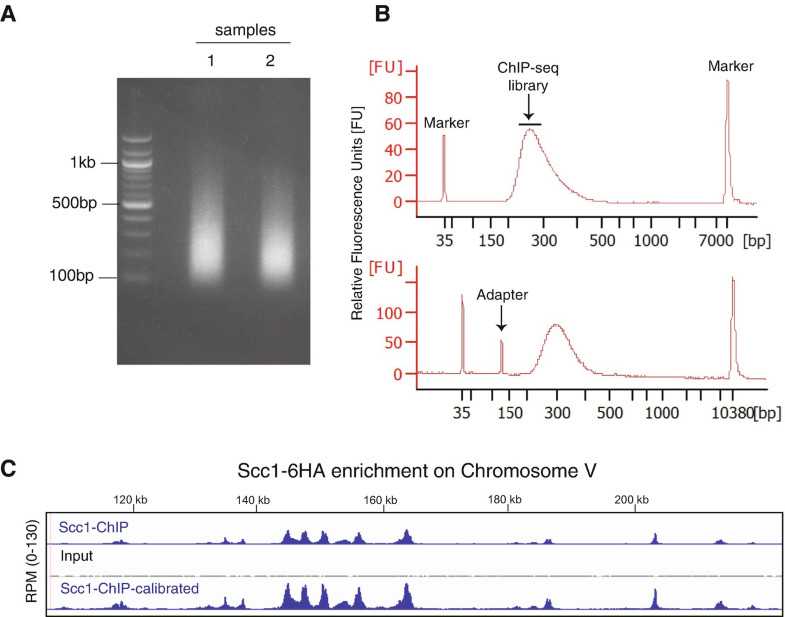
Libraries are now ready for sequencing using a sequencing platform of choice.
The final concentration of the library to load on the flow cell is 1.5 pM with an Input–IP ratio 15%:85%. Perform paired-end sequencing with 76 bp—76 bp for Read 1 and Read 2.

