CUTAC for FFPEs
Steven Henikoff
Abstract
For more than a century, Formalin Fixed Paraffin Embedded (FFPE) sample preparation has been the preferred method for long-term preservation of biological material. However, the use of FFPE samples for epigenomic studies has been difficult because of chromatin damage from long exposure to high concentrations of formaldehyde. Previously, we introduced Cleavage Under Targeted Accessible Chromatin (CUTAC), an antibody-targeted chromatin accessibility mapping protocol based on CUT&Tag. Here we show that simple modifications of our single-tube CUTAC protocol are sufficient to produce high-resolution maps of paused RNA Polymerase II (RNAPII) at enhancers and promoters using FFPE samples. We find that transcriptional regulatory element differences produced by FFPE-CUTAC distinguish between mouse brain tumors and identify and map regulatory element markers with high confidence and precision, including microRNAs not detectable by RNA-seq. Our simple workflows make possible affordable epigenomic profiling of archived biological samples for biomarker identification, clinical applications and retrospective studies.Version 2 includes improvements in both the on-slide and bead options and extension of the improved bead option to curls.

Before start
Steps
REAGENT SETUP (for up to 16 samples)
Cross-link reversal buffer Mix 8 ml 1 M Tris-HCl pH8.0, 2 ml dH2O and 4 µl 0.5 mM EDTA.
Rinse buffer (Option 1) Mix 1 mL 1 M HEPES pH 7.5 and 1.5 mL 5 M NaCl, and bring the final volume to 50 mL with dH2O.
Triton-Wash buffer Mix 1 mL 1 M HEPES pH 7.5, 1.5 mL 5 M NaCl, 250 µl 10% Triton-X100, 12.5 μl 2 M spermidine and 20 µl 0.5 M EDTA, bring the final volume to 50 mL with dH2O, and add 1 Roche Complete Protease Inhibitor EDTA-Free tablet. Store the buffer at 4 °C for up to 2 days.
Primary antibody solution Mix 17 µl RNA Polymerase II-Ser5p: (Cell Signaling Technologies (D9N5I) mAb #13523) + 423 µl Triton-Wash buffer (1:25).
Secondary antibody solution Mix 17 µl guinea pig anti-rabbit (Antibodies Online) with 423 µL Triton-Wash buffer (1:25).
Protein A(G)-Tn5 solution Mix 21 µl Protein A(G)-Tn5 (Epicypher cat. no. 15-1117) with 419 µL Triton-Wash buffer (1:20).
CUTAC-DMF Tagmentation buffer Mix 17.7 mL dH2O, 4 mL N,N-dimethylformamide, 220 µl 1 M TAPS pH 8.5, and 110 µl 1 M MgCl2 (10 mM TAPS, 5 mM MgCl2, 20% DMF). Store the buffer at 4 °C for up to 1 week.
TAPS wash buffer Mix 1 mL dH2O, 10 µl 1 M TAPS pH 8.5, 0.4 µl 0.5 M EDTA (10 mM TAPS, 0.2 mM EDTA). Store at room temperature.
1% SDS/ProtK Release solution (For 16 samples) Mix 10 µl 10% SDS and 1 µl 1 M TAPS pH 8.5 in 79 µl dH2O. Just before use add 10 µL Thermolabile Proteinase K (NEB cat. no. P8111S).
6% Triton Mix 600 µl 10% Triton-X100 + 400 µl dH2O. Store at room temperature.
Option 1: On-slide FFPE-CUTAC deparaffinization in hot cross-link reversal buffer.
Place slides in cross-link reversal buffer in a slide holder that is filled to completely cover the slides. Place the holder in a water bath at 85-90 oC and incubate for at least an hour. The paraffin will melt and float to the top. Remove slide holder to an ice-cold water bath to chill. Adding more solution to overfill will drain off any solid paraffin.
Remove slides to Rinse Buffer in a slide holder.
For Option 1 (on-slide), continue with Step 5. For Option 2 (Magnetic Beads), skip to Step 22.
Option 1 (continued): On-slide FFPE-CUTAC Incubation with primary antibody.
For each slide, remove from slide holder, wick off excess liquid from the glass surface with a Kimwipe (without touching the tissue) and place tissue-side up on a dark surface for visibility. Carefully pipette ~50 µl primary antibody solution over the tissue.
Cover the clear portion of the slide with a rectangle of plastic film (or a square for small tissue sections) using surface tension to spread the liquid, while excluding large bubbles and wrinkles. Place wrapped slides separated in a dry slide holder ( Figure 2 ) or in the rack of a staining dish, which can be used as a "moist chamber" ( Figure 3 ).


Incubate at room temperature for at least 1 hr.
Remove plastic wrap and gently rinse slide by pipetting 1 mL Triton-Wash buffer dropwise over the top of the slide.
Option 1 (continued): Incubation with secondary antibody ( 1.5 hr).
Wick off excess liquid with a Kimwipe and place tissue-side up on a dark surface. Carefully pipette ~50 µl secondary antibody solution over the tissue.
Cover the clear portion of the slide with a rectangle of plastic film using surface tension to spread the liquid, while omitting bubbles and folds. Place wrapped slides separated in a dry slide holder.
Incubate at room temperature for at least 1 hr.
Remove plastic wrap and gently rinse slide 1-2 times with 1 mL Triton-Wash buffer.
Option 1 (continued): Binding Protein A(G)-Tn5 adapter complex (1.5 hr)
Remove from slide holder and wick off excess liquid with a Kimwipe. Place tissue-side up on a dark surface. Carefully pipette ~50 µl pA(G)-Tn5 solution over the tissue.
Cover the clear portion of the slide with a rectangle of plastic film using surface tension to spread the liquid, while omitting bubbles and folds.
Incubate at room temperature for at least 1 hr.
Remove plastic wrap and gently rinse slide 1-2 times with 1 mL Triton-Wash buffer. Drain on paper towel or Kimwipe and place in a slide holder filled with Triton-Wash buffer for 10 min. Drain and place in a slide holder with Triton-Wash buffer for 10 min.
Drain on paper towel and wick off excess liquid with a Kimwipe and place in a slide holder filled with 10 mM TAPS pH 8.5 for 10 min.
Option 1 (continued): Tagmentation and dissection (1.5 hr)
Remove slides and drain on paper towel or Kimwipe and place in a slide holder containing cold Tagmentation buffer.
Incubate 1 hr in a water bath at 55°C.
Remove each slide to a slide holder containing 10 mM TAPS pH 8.5 to hold.
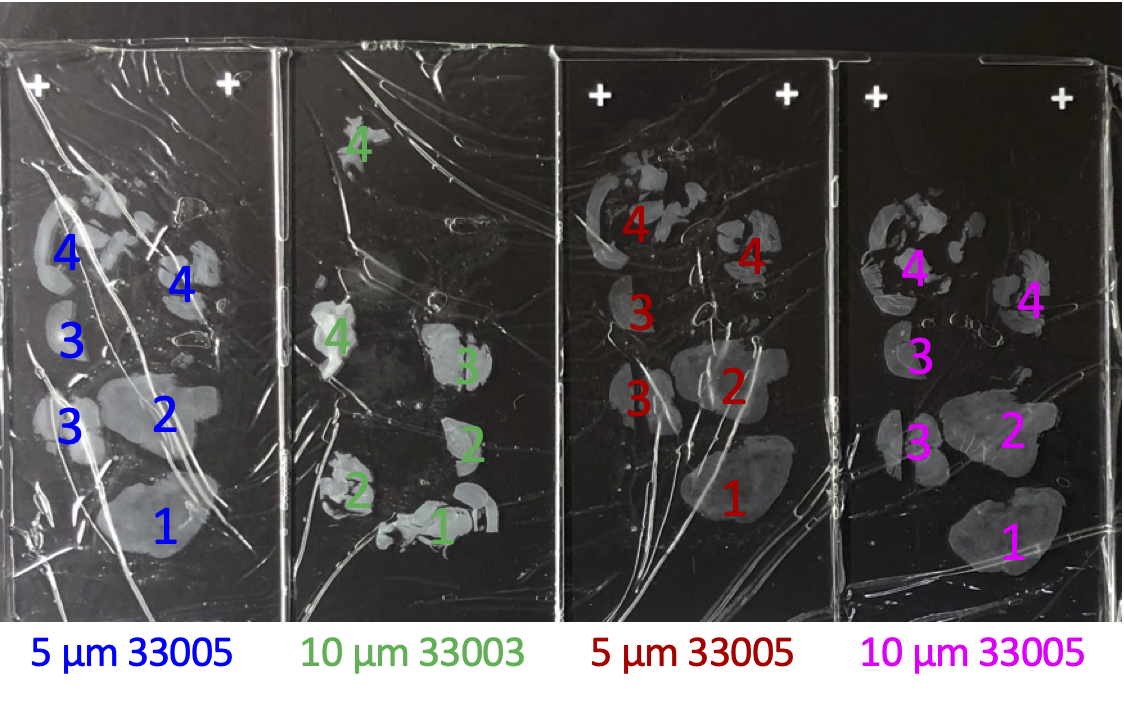
Remove slide from slide holder, drain and use a Kimwipe to remove excess liquid from the top surface. Dissect or scrape using a total of no more than 5 µL 1% SDS/Thermolabile Proteinase K solution per PCR tube. For larger tissue amounts, use more SDS/TLProtK solution and divide up into PCR tubes such that no more than 5 µL is deposited into each tube. To recover all tissue from the slide dice and scrape with a safety razor blade. Vortex and centrifuge to compact beads in the bottom of the tube and proceed to Fragment Release (Step 52).
Option 2: FFPE-CUTAC using beads: Deparaffinization in mineral oil and cross-link reversal buffer.
FFPE slide or curl: Scrape all or part of a 10 µm FFPE slide ( Figures 2-4 ) or a "curl" ( Figure 5 ) into a 1.7 ml tube (e.g. MCT-175-C), add 200 µL mineral oil. Vortex, spin, and place in a 90°C water bath for 5 min. While still warm vortex to fully suspend the paraffin and spin on full.

Using a blue pestle attached to a pestle motor place the pestle into the bottom of the tube, start the motor and homogenize with short up-and-down motions for ~20 sec.
Add 200 µl warm Cross-link reversal buffer, then 6 µl 1:10-diluted Biomag Plus amine beads into the bottom (aqueous) layer, vortex and homogenize ~20 sec with the motorized pestle.
Add 800 µl warm Cross-link reversal buffer and vortex to mix, spin on full and replace in the 90°C water bath. Incubate ≥1 hr.
Remove from water bath, mix by hard vortexing and spin on full. Very carefully remove the top (oil) layer without disturbing the interface, where there will be trapped tissue and beads, leaving behind a thin oil layer above the meniscus.
Add 500 µl mineral oil, mix by inversion (do not vortex), spin on full and carefully remove the mineral oil layer leaving behind a thin oil layer. Repeat with a second 500 µl mineral oil wash. Respin to clear tube sides and pipet off excess oil, leaving behind a thin oil layer above the meniscus.
Add 2.4 µL (undiluted) Pierce glutathione beads to bottom of tube avoiding the mineral oil on the surface. Mix by inversion.
Do a quick spin and place on the magnet stand. When clear carefully remove the supernatant using a 200 µL low-bind pipette tip.
Add 1 ml Triton-Wash buffer and vortex followed by a quick spin, and divide into two or more PCR tubes. The following assumes two PCR tubes per scrape or curl, one for RNAPII-Ser5p and one for H3K27ac, but for smaller aliquots volumes should be adjusted to maintain the concentration of reagents.
Place tubes on magnet stand and carefully remove supernatant using a low-bind 200 µL pipette tip.
Option 2 (continued): Incubation with primary antibody
Resuspend beads in 100 µl primary antibody solution followed by vortexing.
Incubate at least 1 hr on Rotator or Nutator at room temperature.
Option 2 (continued): Incubation with secondary antibody
After a quick spin, place the tubes on the magnet stand to clear and withdraw and discard the antibody supernatant using a 200 µL low-bind pipette tip.
Resuspend beads in 100 µl secondary antibody solution followed by vortexing.
Incubate at least 1 hr on Rotator or Nutator at room temperature.
After a quick spin, place the tubes on the magnet stand and withdraw and discard the antibody supernatant using a 200 µL low-bind pipette tip.
While on the magnet stand, slowly drip in 500 µl of Triton-Wash buffer. Carefully withdraw and discard the wash supernatant using a 200 µL low-bind pipette tip. Proceed immediately to the next step.
Option 2 (continued): Binding Protein A(G)-Tn5 adapter complex
Mix pAG-Tn5 pre-loaded adapter complex in Triton-Wash buffer following the manufacturer's instructions (e.g. 1:20 for EpiCypher pAG-Tn5).
Add 100 µl pA(G)-Tn5 mix followed by vortexing. Place the tubes on a Rotator or Nutator at room temperature for ≥1 hr.
After a quick spin, place the tubes on the magnet stand and withdraw and discard the pA(G)-Tn5 supernatant using a 200 µl low-bind pipette tip.
While on the magnet stand, slowly drip in 500 µl of Triton-Wash buffer. Carefully withdraw and discard the wash supernatant using a 200 µl low-bind pipette tip.
While on the magnet stand, add 200 µl TAPS wash. Withdraw and discard the TAPS wash supernatant using a 200 µL low-bind pipette tip. Proceed immediately to the next step.
Option 2 (continued): Tagmentation
Resuspend the bead/FFPE pellet in 100 µl CUTAC-DMF tagmentation solution (5 mM MgCl2, 10 mM TAPS, 20% DMF, 0.05% Triton-X100) while vortexing. Incubate at 55°C for 1 hr in a thermocycler.
After a quick full centrifugation, place the tubes on a magnet stand and withdraw and discard the Tagmentation buffer supernatant using a 200 µl low-bind pipette tip.
While on the magnet stand, add 100 µl TAPS wash. Withdraw and discard the TAPS wash supernatant using a 200 µl low-bind pipette tip.
Add 10 µl 1% SDS/Thermolabile Proteinase K solution per PCR tube. Vortex, quick spin and proceed to Fragment Release (Step 48).
Fragment Release and PCR
Incubate at 37ºC for 30 min and 58ºC for 30 min (programmed in succession in a PCR cycler with a heated lid) to release pA-Tn5 from the tagmented DNA. Open the tubes and add 15 µL 6% Triton-X100, close and incubate at 37ºC for 30 min on the cycler.
Add 2 µl of 10 µM Universal or barcoded i5 primer + 2 µl of 10 µM barcoded i7 primers, using a different barcode pair for each sample. Vortex on full and place tubes in the metal tube holder on ice.
Add 25 µl NEBnext (non-hot-start), vortex to mix, and perform a quick spin. Place the tubes in the thermocycler and proceed immediately with the PCR.
Begin the cycling program with a heated lid on the thermocycler:
Cycle 1: 58°C for 5 min (gap filling)
Cycle 2: 72°C for 5 min (gap filling)
Cycle 3: 98°C for 5 min
Cycle 4: 98°C for 10 sec
Cycle 5: 63°C for 30 sec
Cycle 6: 72°C for 1 min
Repeat Cycles 4-6 11 times
Hold at 8 °C
Post-PCR Clean-up (30 min)
After the PCR program ends, remove tubes from the thermocycler, vortex to resuspend, and add 130 µL of SPRI beads (ratio of 1.3 µL of SPRI beads to 1 µL of PCR product). Mix by pipetting up and down.
Let sit at room temperature 5-10 min.
Place on the magnet stand for a few minutes to allow the solution to clear.
Remove and discard the supernatant.
Keeping the tubes in the magnet stand, add 400 µL of 80% ethanol.
Completely remove and discard the supernatant.
Repeat Steps 56 and 57.
Perform a quick spin and remove the remaining supernatant, avoiding air drying the beads by proceeding immediately to the next step.
Remove from the magnet stand, add 22 µl 10 mM Tris-HCl pH 8, vortex and quick spin. Let sit for at least 5 min to elute the DNA.
Place on the magnet stand and allow to clear.
Remove the liquid to a fresh 1.5 mL tube with a pipette, avoiding transfer of beads.
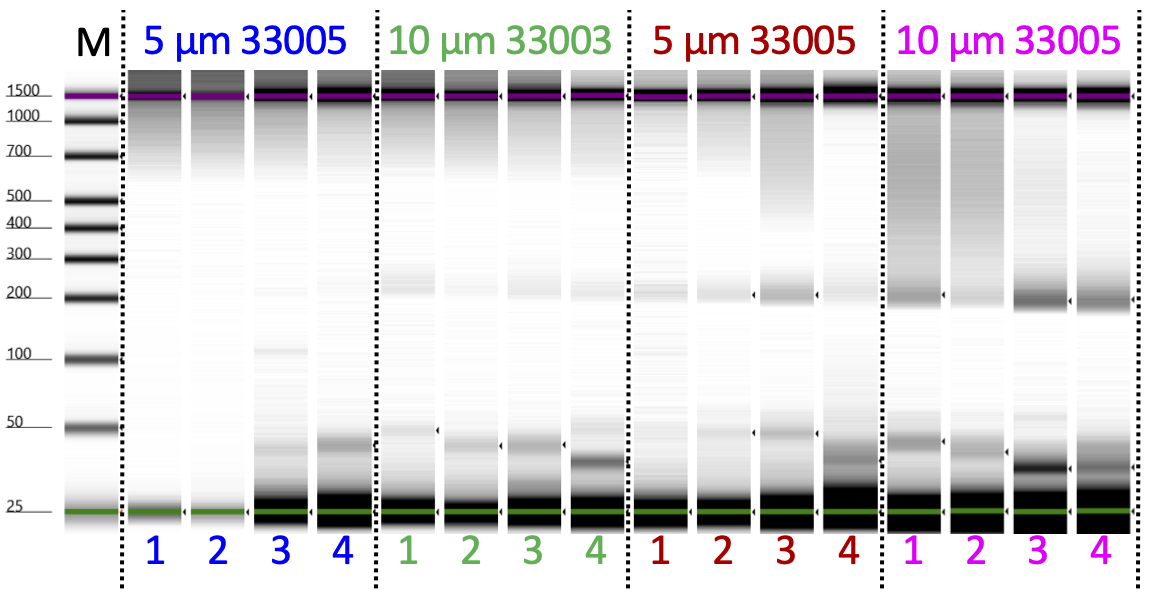
Tapestation analysis and DNA sequencing
Determine the size distribution and concentration of libraries by capillary electrophoresis using an Agilent 4200 TapeStation with D1000 reagents or equivalent.
Mix barcoded libraries to achieve equal representation as desired aiming for a final concentration as recommended by the manufacturer. After mixing, perform an SPRI bead cleanup if needed to remove any residual PCR primers.
Perform paired-end Illumina sequencing on the barcoded libraries following the manufacturer’s instructions.
Data processing and analysis
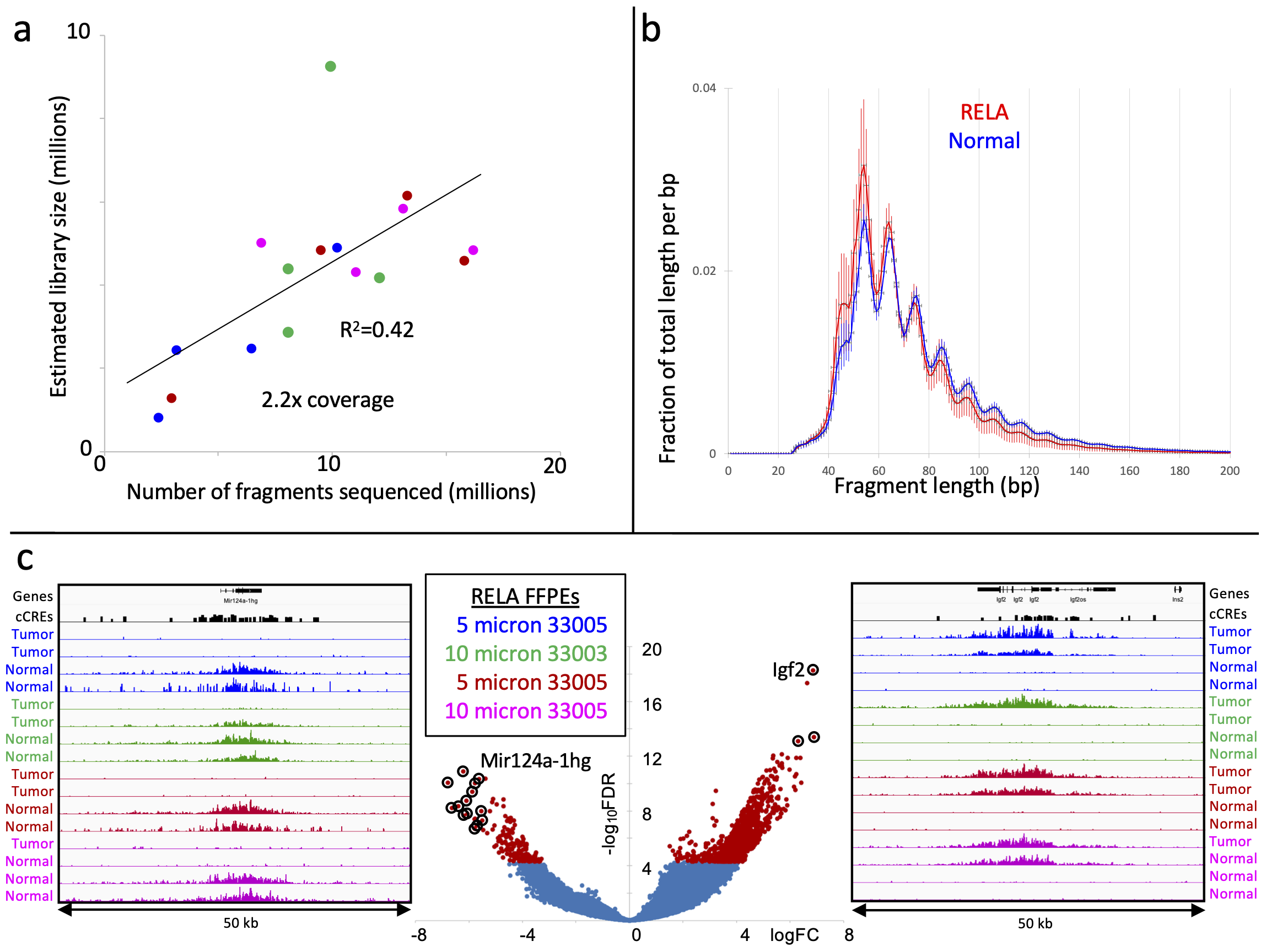
Align paired-end reads to hg19 using Bowtie2 version 2.3.4.3 with options: --end-to-end --very-sensitive --no-unal --no-mixed --no-discordant --phred33 -I 10 -X 700. For mapping E. coli carry-over fragments, we also use the --no-overlap --no-dovetail options to avoid possible cross-mapping of the experimental genome to that of the carry-over E. coli DNA that is used for calibration. Tracks are made as bedgraph files of normalized counts, which are the fraction of total counts at each basepair scaled by the size of the hg19 genome.
Our CUT&Tag Data Processing and Analysis Tutorial on Protocols.io provides step-by-step guidance for mapping and analysis of CUT&Tag sequencing data. Most data analysis tools used for ChIP-seq data, such as bedtools, Picard and deepTools, can be used on CUT&Tag data. Analysis tools designed specifically for CUT&RUN/Tag data include the SEACR peak caller also available as a public web server and CUT&RUNTools.