Bioluminescence-based Minimum Inhibitory Concentration (MIC) testing of fungal extracts against Escherichia coli
Siouxsie Wiles, Shara Van De Pas
Abstract
In this protocol, we describe how to obtain the minimum inhibitory concentration (MIC) of fungal extracts using a bioluminescent derivative of Escherichia coli ATCC 25922.
Before start
Prepare media. You will also need an overnight culture of E. coli to test against. The day before, Inoculate 10 mL of MHB in a 50 mL tube with E. coli 25922 lux and incubate overnight at 37 degrees C with shaking at 200 rpm.
Steps
Preparing 96-well plates
We test doubling dilutions of each extract fraction in duplicate with a maximum concentration of 1mg/mL. Each round of screening also requires a control plate containing the solvent the extract was dissolved in (e.g. DMSO), an antibiotic (to be used as a positive control, e.g. erythromycin), and broth (negative control to test the growth of the testing organism). Using the plate layout described in Figure 1, each plate can contain either one complete set of a crude extract and five fractions of decreasing polarity, or the appropriate controls for the testing round.
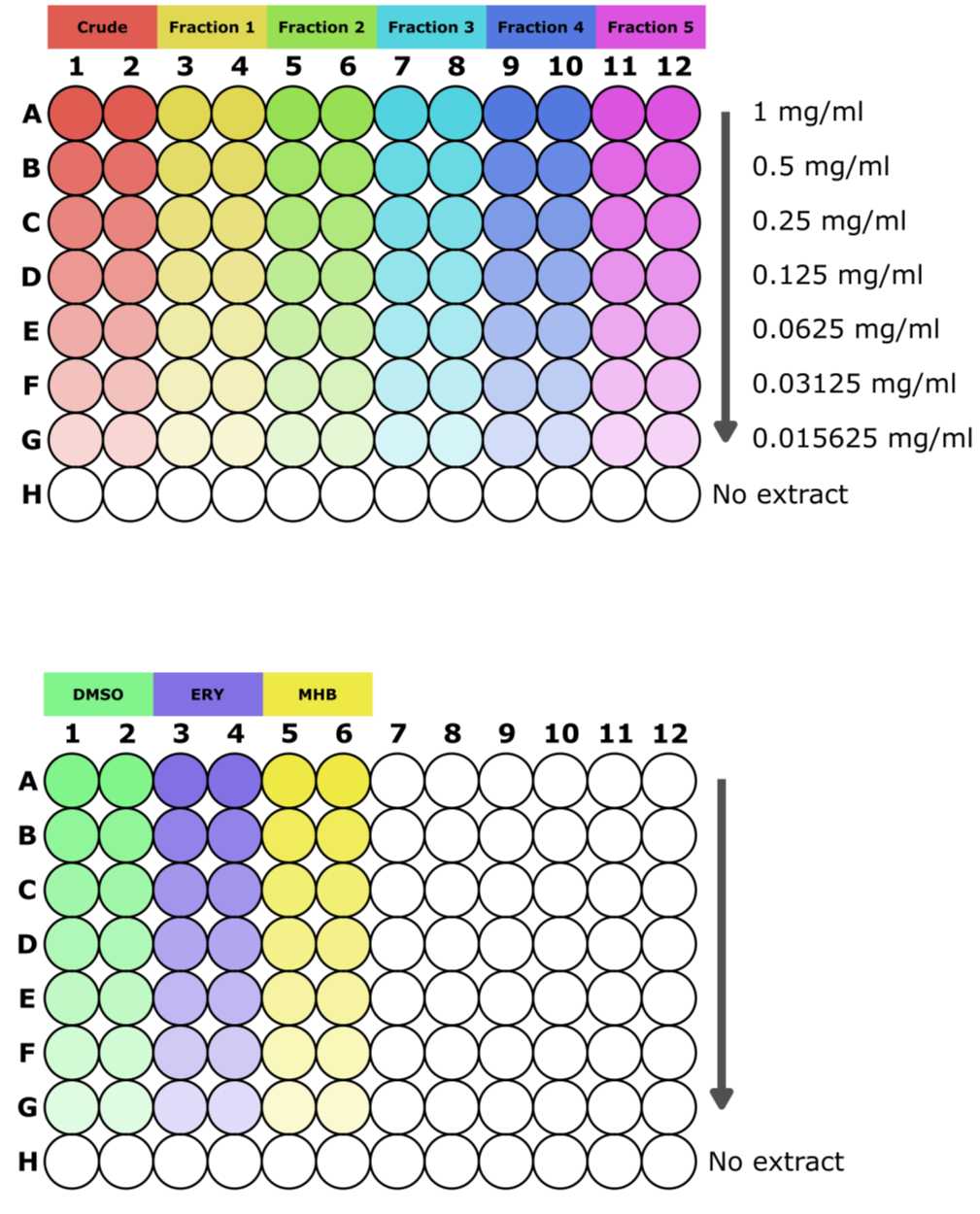
Set up your plate according to Figure 1. Use masking tape to make a front label on the lid including your name or initials, the date, the name of the bacteria you are testing against, and the name of the extract being tested.
The final volume for each well is 100µL . Add 50µL of Mueller Hinton Broth (MHB) to all the wells on the first plate except for the top row (A).
Add all the extracts at double their required concentration to row A of the plate. If the extracts were dissolved at 50mg/mL , add 96µL MHB to row A and 4µL of each extract fraction. For the controls add 96µL MHB to row A in the "DMSO" columns and 4µL DMSO to these columns. Add 1µLof 50mg/mL Erythromycin to the "ERY columns" and 99µLMHB. Lastly, add 100µL sterile MHB to the "Broth columns."
Using a multichannel pipette, gently aspirate repeatedly to homogenise the wells of each row. Then transfer 50µLfrom the first row to the second row and aspirate to mix. Discard tips and repeat the doubling dilution down the plate, changing tips between rows, until you reach row G. Do not continue the dilution into row H.
Aspirate 50µL from the wells in row G and discard the solution. This will leave row H as a growth control containing no extract/DMSO/antibiotic.
Preparing bacterial inoculum
This step needs to be done the day before. Inoculate 10mL of MHB in a 50 mL tube with E. coli 25922 lux and incubate overnight at 37°C with shaking at 200 rpm.
Measure the optical density of the overnight culture of E. coli at 600nm (OD600). We do this by diluting a sample of the overnight culture 1:10 in a 1.5mL cuvette with MHB ( 720µL broth + 80µL bacteria).
Dilute the bacterial culture with MHB to give a final OD600 of 0.001 which is the equivalent of ~5 x 105 bacteria per mL. We do this by diluting the bacterial culture to an OD600 of 1 and then diluting that 1 in 1000 to give 0.001.
Make up the bacterial inoculum in a 50ml Falcon tube using a serological pipette to add the appropriate volume of MHB. Tip the inoculum into a pipetting reservoir and use a multichannel pipette to add 50µL to all the wells excluding the sterile MHB control columns.
Checking inoculum concentration
Add 90µL of MHB or Phosphate Buffer Solution (PBS) to each of the wells in a single column of a clear 96-well plate. Mix in 10µL of bacterial inoculum to the top well, mix and discard the tip. Using a clean tip, remove 10µL, add it to the next well in the column and mix. Repeat to perform a 10-fold serial dilution down the column, using a clean tip each time.
Plate three technical replicates of each dilution onto a MHA plate. Incubate agar plates upside down at 37 °C overnight. Count the colonies the following day.
Count visible colonies to ensure theinoculum was correct. It should be approximately ~5 x 105CFU/mL
Measuring bioluminescence
We use a Perkin Elmer Victor X plate luminometer set to read 96 well plates with an integration time of 1 second per well. If we haven’t filled the entire plate, we change the settings so that the machine doesn’t measure the empty wells.
We take measurements immediately after setting up the plate (t=0) and then at 2, 4, 6, and 24 hours.
Between measurements, place lids on the plates, put them in a plastic box lined with damp paper towels, and incubate at 37 °C with shaking at 100 RPM.
After the final timepoint, if the light has reduced to background levels in any of the wells (for our machine this is < 10 relative light units [RLU]) plate 3 x 10 µL aliquots from each "dark" well onto fresh MHA to check for bacterial viability. We define the minimum bactericidal concentration (MBC) as the lowest concentration at which no colonies appear after overnight incubation at 37 °C.

