Bacterial 16S-V4V5 rRNA amplification for NGS Illumina sequencing (Metabarcoding)
Estelle Bigeard, Julie Dinasquet
gene amplification
metabarcoding
NGS
sequencing
bacteria
archea
16rRNA gene
ribosomal rna sequencing
Abstract
For metabarcoding purpose, the first step involves the amplification by PCR of a given gene region (for example V3 or V5 region of 16S rRNA gene) or gene itself if its size does not exceed 600bp (the longest fragment size that can be sequenced by Illumina technology).
The defined forward and reverse primers that are complementary upstream and downstream of the region of interest, needs to be designed with overhang adapters which will be used in a subsequent limited‐cycle amplification step, in order to add the dual‐index barcodes and Illumina flow cell adapters. To design illumina primers, it will be necessary to know the sequencing method, and therefore the adapters sequence.
The following protocol explains the amplification of variable regions 4 and 5 of the 16S rRNA gene for prokaryote metabarcoding analyses from seawater samples for sequencing by the Get-PlaGE platform, Toulouse, France.
The 515f-806r bacterial/archaeal primer pair, designed by Caporaso et al. (2011), was shown to be biased against some aquatic bacteria. The alternative primer set modified with additional degeneracy (Parada et al. (2015) and Apprill et al. (2015)) reduce the biais against important marine prokaryotes (such as the SAR11 clade, Crenarchaeota/Thaumarchaeota).
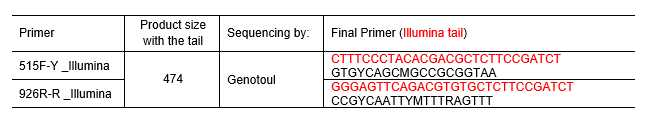
Primers 515F-Y (5'-GTGYCAGCMGCCGCGGTAA) and 926R-R (5'-CCGYCAATTYMTTTRAGTTT) yield more estimates of mock community abundances and produce longer amplicons.
Steps
Samples : Acid nucleic extraction and quantification
Extract DNA/RNA from the samples using the method/kit appropriate to the specific samples.
When working with RNA, another step to transform the RNA in cDNA is necessary.
Quantify and quality-check the final DNA via NanoDrop or Qubit/PicoGreen Method.
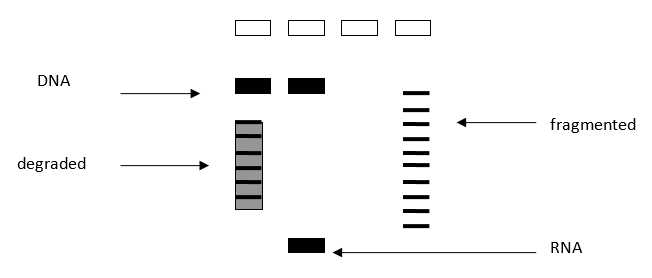
The A280/260 ratios should be 1.8 or higher and once working with total community extracted DNAs, a concentration of at least 1 ng/μLis desirable to get consistent PCR results. Avoid secondary extractions or clean-ups for inhibitors, until PCRs have truly shown inhibition (example after trying the PCR by diluting the original DNA and/or adding BSA to the final PCR reaction)–many suboptimal DNAs will still work in PCR. A gel can be run to verify integrity of the extracted material, but it is generally unnecessary for PCR-only studies ( Figure 1 ). Some protocols suggest the normalization of the DNA samples concentration prior to the PCR. In our experience, this is not always possible given the low concentration of some samples. The volume of sample added to the PCR can be settled in function of a range of concentrations instead of normalizing all the samples to the same concentration.
Load in agarose gel 0.8%, 1μl DNA + loading buffer. The limit of DNA detection using EtBr gels is around 10ng.
Each DNA sample have to be normalized to 5ng/µl for PCR. For that, dilute DNA in nuclease free water.
It is not necessary to check the DNA concentration after dilution.
Samples with a DNA concentration less than 5ng/µl will be amplified as is in PCR.
PCR amplification
Below is a suggestion of protocol for bacterial 16S amplification from total genomic DNA. This protocol has been tested and used with success.
However, before starting the library preparation, few samples from the batch (es) to be analyzed should be selected and the PCR conditions should be tested. The tests can be performed by using the primers without the illumina “tail” which are less expensive since it can be order by normal purification method, like Se-POP. By using also these primers, it avoids excessive manipulation of the primers with the illumina tail and therefore the chances of contamination. Low number of cycles, pool multiple (i.e., triplicate) PCRs for each sample, high initial time of denaturation are fundamental practices for the PCR when targeting diversity studies and should be considered for all reactions, including the tests in order to reduce or avoid PCR bias caused by PCR selection and PCR drift.
• PCR selection includes all mechanisms which inherently favor the amplification of certain templates due to properties of the genes and/or of their flanking sequences of the target region. Potentially important contributors to PCR selection are preferential denaturation due to overall low GC content, higher binding efficiency of GC-rich permutations of degenerate primers, differential accessibility of rRNA genes within genomes, and correlation between amplification probabilities and gene copy numbers within genomes.
• PCR drift is cause by stochastic variation in the early cycles of the reaction (when amplification still proceeds largely from the genomic templates), and its outcome should therefore not be reproducible in replicate PCR amplifications.
More details about these biases can be found in the following references:
-Aird, D. et al. Analyzing and minimizing PCR amplification bias in Illumina sequencing libraries. Genome Biol. 12, R18 (2011).
-Wagner, A. et al. Surveys of Gene Families Using Polymerase Chain Reaction: PCR Selection and PCR Drift. Syst. Biol. 43, 250 (1994).-Polz, M. F. & Cavanaugh, C. M. Bias in template-to-product ratios in multitemplate PCR. Appl. Environ. Microbiol. 64, 3724–3730 (1998).
Primers used for DNA amplification and sequencing
Use the existing primers (Table 1 & Table 2) or design your own custom gene primers with the proper Illumina indices and Index adaptor orientations. These adapters will be different in function of the libraries preparation protocol, and therefore in function of sequencing platform (Table 2).
It is recommended to order the illumina primers with a good synthesis and purity parameters.
We have ordered at EUROGENTEC the NGS primers with RP-HPLC purity method, 10 or 40nmol synthesis scale and at 100µM TE concentration. We could also order in dried and dissolve them with nuclease free water or TE 1x sterile buffer to obtain 100µM stock solution (as described on the Eurogentec’s technical data sheet).
Then we dilute them to 10 μM working concentration (1/10th the typical 100 μM working stock concentration for primers). This proceed should be performed in the PCR hood and with molecular grade water.
The primers (stock and working solutions) are stored at -20°C.

Controles
Test a negative control for each series. The negative PCR product could be inserted in the library to have a control of air contaminations.
It is possible to test a positive control (mock communities) to check the sequencing efficiency and sequencing error. For that, you have to select axenic cultures, to know the starting cells concentration and the DNA concentration. We could buy commercial Mock communities (ex: Zymo).
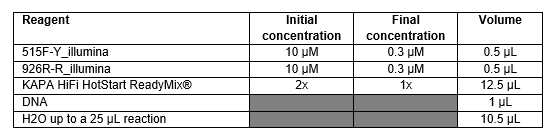
Amplification with Kapa HotStart ReadyMix® (1 round)
Place plates, tubes and nuclease free waterto prepare the PCR mix under UV .
It is best to use an enzyme no derived from bacteria, and to limit the number of amplification cycles.
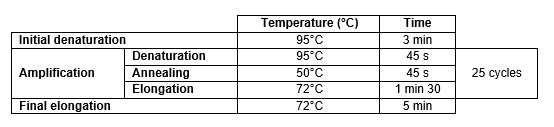
PCR are performed as follows (in triplicate):
Gel electrophoresis
PCR products should be checked initially by gel electrophoresis for unspecific amplification and band size.
- Check the PCR products on agarose gel to see if the bands are weak or strong.
Prepare an agarose gel 1-1.5% in 0.5X TAE buffer
Microwave until the solution is clear (2 minutes)
Stain with 20 µL ethidium bromide per 100ml (or SYBR-safe dye)
Prepare tray & comb(s)
Pour gel into tray
When the gel is polymerized, remove comb
Load gel in electrophesis cuve containing the same 0.5X TAE buffer
Gel must be covered
Cut one strip of parafilm.
Load 1-2 µL loading dye onto parafilm, one drop per sample
Mix 1 to 3 μl of each PCR product with loading dye (up & down 3 times)
Load it in the agarose gel
Load 3 µL of SmartLadder into first and/or last well.
Close & Plug in electrodes to power source
Run the migration for ~120 V, 30-45 minutes
Observe results using a UV imager like Image Quant LAZ4000, Ge Healthcare
The negative control must be negative and the positive control must be positive to validate the PCR.
The size of amplicons is approximatly 400 bp.
It is possible to obtain some bands of longer size.
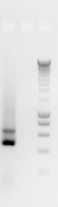
- Samples with failed PCRs (or spurious bands) are re-amplified by optimizing the PCR (further template dilution to 1:100 or using BSA/other additives) to produce correct bands, or purified using a purification kit like “Nuclesopin Gel & Clean up kit– Macherey-Nagel”.
Amplicons are stored at -20°C until treatment.
PCR Purification
This step allows to pool triplicates and clean PCR products, in particular the band of interest.
For that, we use the purification kit from Macherey-Nagel™ : NucleoSpin™ Gel and PCR Clean-up Kit.
2 methods:
-
Clean PCR products in solution
-
Load the PCR products in totality into a gel , cut the band of interest and clean the PCR products in gel agarose.
Follow manual instructions, Instruction-NucleoSpin-Gel-and-PCR-Clean-up.pdf (mn-net.com), with severals modifications:
-
First elution : add 30µl TE preheated at 70°C onto the column, incubate at 70°C for 5 minutes
-
Centrifuge at 11000g 1 min
-
Second elution: add 30µl TE preheated at 70°C or the previous eluate onto the column
-
Centrifuge at 11000g 1 min
-
Aliquote 2.5µl for the quantification
The purified PCR products are stored at -20°C.
Control of the quantity & quality
Purified PCR products are measured using Nanoquant Plate and Spark Tecan.
This method allow to measure the concentration of amplified products and the 260/280 and 260/230 ratios using only 2µl of PCR product.
A more precise concentration could be measured using Qubit 4 fluorometer.
If the PCR’s product are not concentrated enough, you could purifie and concentrate them in a smaller volume using a purification and concentration kit like “Nuclesopin Gel & Clean up kit– Macherey-Nagel”.
Preparation of libraries for Illumina sequencing (metabarcoding)
Send to the sequencing platform pictures of gel to check they are conform (purity, degradation, bands intensity, etc) and follow its protocol.
Once PCR products are correct, they are organized in appropriated PCR plates to send (dryice) to the sequencing platform and generate libraries.
We use a high throughput sequencing using paired-end 2x250bp Illumina MiSeq.
If the illumina sequencing is realized on the Genomer Platform in Roscoff (France), see the protocols.io "Preparation of libraries (Metabarcoding) for Illumina sequencing - Genomer Platform".


