BIND&MODIFY: Long-range single-molecule mapping of chromatin modification in eukaryotes
Chen Zhang, Zhe Weng, Fengying Ruan, Weitian Chen, Zhichao Chen, Yeming Xie, Zhe Xie, Juan Wang, Yuxin Sun, Yitong Fang, Mei Guo, Chong Tang, Yiqin Tong, Yaning Li
Abstract
Here we describe a powerful method, BIND&MODIFY, for probing histone modifications and transcription factors at single molecular level. Our approach used the recombinant fused protein A-M.EcoGII, which tethers the methyltransferase M.EcoGII to the protein binding sites and locally labels the neighboring DNA regions via artificial methylations. This method could reveal ingle-molecule heterogenous histone modification status and CpG methylation at the same time, and could enable quantify the correlation between the distal elements. Further applications based on this method's concept could be applied to probe multiple protein binding events on the same single molecular DNA. The method proposed herein may soon become an essential tool for third-generation sequencing in the future.
Steps
OVERVIEW
This protocol describes step-by-step guidelines for BIND&MODIFY method.BIND&MODIFY method is based on the indirect labelling of DNA regions bound to the protein of interest (with antibody) using an engineered recombinant fusion protein, protein A-M.EcoGII (pA-M.EcoGII), whose methyltransferase activity can be locally controlled.Firstly, BIND&MODIFY method shows comparable distribution of histone modifications (H3K27me3) and DNA binding protein (CTCF) with conventional ChIP-seq method. Secondly, BIND&MODIFY method resolves histone modification in complex genomic region, phases the epigenome, and uncovers epigenomic heterogeneity at single molecular level. Furthermore, BIND&MODIFY method reveals long-distance correlation between genome regulators. We believe BIND&MODIFY method to become one powerful tool for probing DNA binding protein and their regulatory mechanisms in the upcoming long-read sequencing technology arsenal.
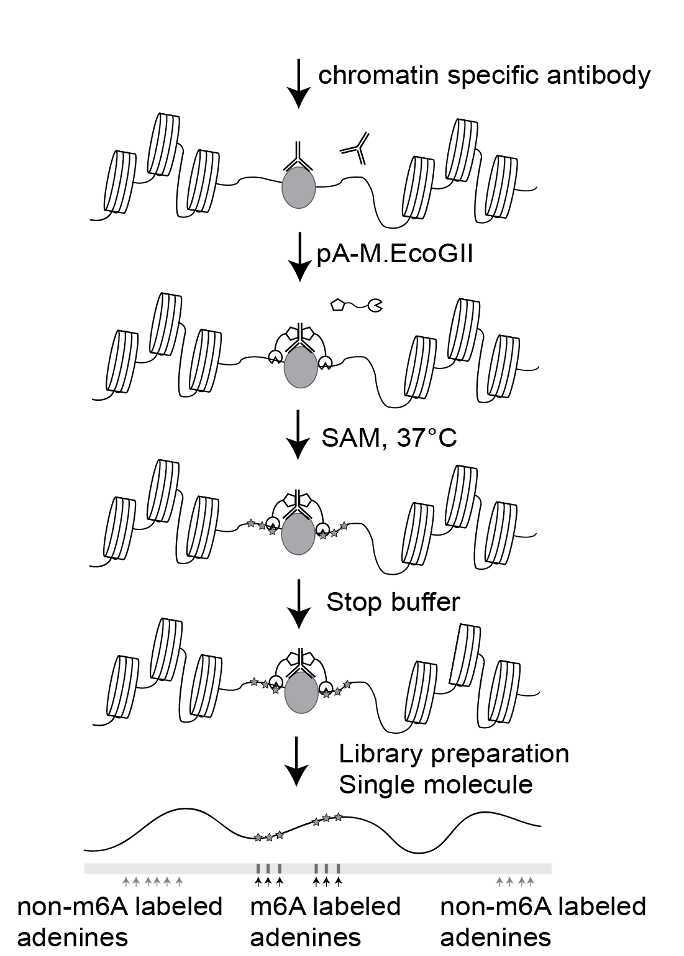
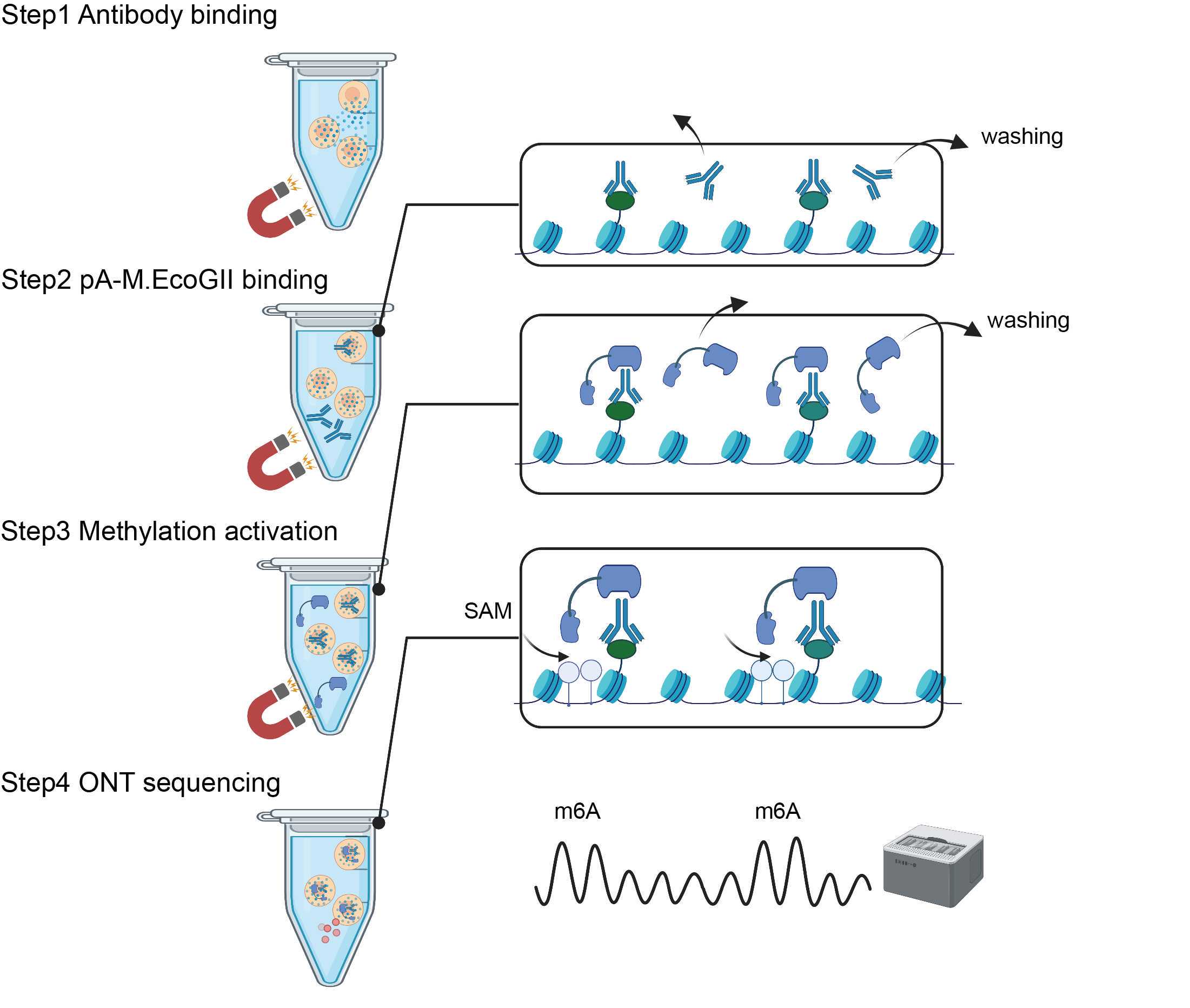
REAGENT SETUP
Digitonin (5%) : Dissolve 100mg digitonin in 2 ml DMSO. Aliquote at 50ul per PCR tube and freeze at -20. Avoid freeze-thaw cycles.
Caution : Digitonin is toxic and avoid any direct contact with skin or during breath. Use full PPE including a mask, lab coat and gloves while handling with digitonin. DMSO can penetrate through the skin.
Binding Buffer : Prepare fresh and store the buffer at 4°C for 6 months.
| A | B | C |
|---|---|---|
| Reagent | Quantity | Final Concentration |
| 1M HEPES pH 7.9 | 200ul | 20mM |
| 1M KCl | 100ul | 10mM |
| 1M CaCl2 | 10ul | 1mM |
| 1M MnCl2 | 10ul | 1mM |
| H2O | 9750ul | |
| Total | 10ml |
Wash Buffer : Prepare fresh, and store the buffer at 4°C up to 1 week.
| A | B | C |
|---|---|---|
| Reagent | Quantity | Final Concentration |
| 1M HEPES pH 7.5 | 1.0ml | 20mM |
| 5M NaCl | 1.5ml | 150mM |
| 2M Spermidine | 12.5ul | 0.5mM |
| Roche Proteinase Inhibitor cocktail tablet | 1 tablet | 1X |
| H2O | 47.5ml | |
| Total | 50ml |
Dig-wash Buffer : Prepare fresh, store the buffer at 4°C up to 2 days.
| A | B | C |
|---|---|---|
| Reagent | Quantity | Final Concentration |
| 5% Digitonin | 400ul | 0.05% |
| 1X Wash Buffer | 39.6ml | |
| Total | 40ml |
Antibody Buffer : Prepare fresh.
| A | B | C |
|---|---|---|
| Reagent | Quantity | Final Concentration |
| 1M HEPES pH 7.5 | 100ul | 20mM |
| 5M NaCl | 150ul | 150mM |
| 2M Spermidine | 1.25ul | 0.5mM |
| 50X Roche Proteinase Inhibitor cocktail | 100ul | 1X |
| 5% Digitonin | 50ul | 0.05% |
| 0.5M EDTA | 20ul | 2mM |
| 20% BSA | 25ul | 0.10% |
| H2O | 4555ul | |
| Total | 5ml |
Methylation Buffer : Prepare fresh, store the buffer at 4°C up to 2 days.
| A | B | C |
|---|---|---|
| Reagent | Quantity | Final Concentration |
| 10X CutSmart Buffer | 30ul | 1x |
| 50X Roche Proteinase Inhibitor cocktail | 6ul | 1x |
| 5% Digitonin | 3ul | 0.05% |
| 32mM SAM | 7.5ul | 800uM |
| 20% BSA | 1.5ul | 0.10% |
| H2O | 252ul | |
| Total | 300ul |
Digestion Buffer : Prepare fresh.
| A | B | C |
|---|---|---|
| Reagent | Quantity | Final Concentration |
| Polyvinylpyrrolidone 40 | 0.1g | 1% |
| Sodium metabisulfite | 0.1g | 1% |
| 5M NaCl | 1.0ml | 0.5M |
| 1M Tris-HCl, pH 8.0 | 1.0ml | 0.5M |
| 0.5M EDTA | 1.0ml | 50mM |
| 20% SDS | 625ul | 1.25% |
| H2O | 6375ul | |
| Total | 10ml |
Mix and incubate at 65°C during at least 30 minutes. The solution need to be clear before use.
Serapure Beads Solution : Store the solution at 4°C for 1 month.4ml serapure beads wash 4 times with water to remove sodium azide,then resuspend in 10ml serapure beads solution.
| A | B | C |
|---|---|---|
| Reagent | Quantity | Final Concentration |
| 50% PEG8000 | 3.6 ml | 18% |
| 5M NaCl | 2 ml | 1M |
| 1M Tris-HCl, pH 8.0 | 0.1ml | 10mM |
| 0.5M EDTA | 20ul | 1mM |
| 100% tween 20 | 5ul | 0.05% |
| H2O | 4275ul | |
| Total | 10ml |
Bead Binding Buffer : Store the solution at 4°C for 1 week.
| A | B | C |
|---|---|---|
| Reagent | Quantity | Final Concentration |
| 50% PEG 8000 | 4ml | 20% |
| 5M NaCl | 6ml | 3.0M |
| Total | 10ml |
Mix until the solution becomes clear. If PEG8000 is not dissolved, it can lead to a poor yield because PEG8000 makes gDNA to bind to the beads.
Bead Washing Buffer : Prepare fresh.
| A | B | C |
|---|---|---|
| Reagent | Quantity | Final Concentration |
| Absolute Ethanol | 35ml | 70% |
| H2O | 15ml | |
| Total | 50ml |
Cells Preparation
Harvest fresh cells at room temperature.
Note: Use Eppendorf DNA LowBind tube during the whole protocol to reduce cell/DNA loss.
Centrifuge cells at 300g for 5 min at 4°C.
Resuspend the cells in 1ml cold PBS, repeat step 5-step 6 twice.
Resuspend the cells in 900ul cold PBS. Count the cells. For lightly fixed cells, go to step 8.
Normally this protocol works for 5x105 cells per methylation reaction. Aliquot 4x106 cells per centrifuge tube for 8 tube methylation reaction.
Add freshly prepared 1% formaldehyde into the resuspended cells (100ul into 900ul cells), and put at room temperature for 10 min.
Stop the crosslinking by adding 1.25M glycine to twice molar ratio of formaldehyde (60ul into 1ml cell fixing reaction).
Centrifuge cells at 500g for 5 min at 4°C. Carefully remove all the liquids from the supernatant with 1000ul pipette tip followed by 100ul and 10ul pipette tip.
Resuspend the cells with Wash Buffer and count the fixed cells.
Normally this protocol works for 5x105 cells per methylation reaction. Aliquot 4x106 cells per centrifuge tube for 8 tube methylation reaction.
Bind cells or nuclei to Concanavalin A-coated beads
Gently vortex and resuspend the ConA beads slurry, 10ul of the ConA beads would be enough for 5x105 cells. The following is for 4 samples.
Aliquot 90ul ConA beads slurry into 1ml Binding Buffer in a 1.5ml tube and mix by pipetting. Put the tube on a magnetic stand to clear (1-2min).
Remove the liquid completely on the magnetic stand. Add 1ml Binding buffer and mix by pipetting. Quick spin the tube to remove the liquid from the cap.
Put the tube on a magnetic stand to clear, remove the liquid, and resuspend in 90ul Binding Buffer (10ul per sample) and place the activated beads slurry at room temperature until cells are prepared.
Carefully add in 90ul Binding Buffer containing ConA beads into the tube containing 4x106 cells prepared from step 7 or 11. Place on end-over-end rotator for 10min.
Bind primary antibody
Quick spin the tube to remove the liquid from the cap. Place the tube on a magnetic stand to clear, remove the liquid.
Resuspend the cells in 400ul ice cold Antibody Buffer with gentle vortexing. Divide into 8 1.5ml Eppendorf LowBind tubes, and 50ul each tube. Scale up or down based on specific applications.
Add 0.5-1.0ul of specific primary antibody (H3K27me3 or CTCF) into each tube with gentle vortexing.
Note : We use 1:50-1:100 primary antibody dilution as recommended by CUT&TAG protocol.
Place the tube on end-over-end rotator at 4°C overnight.
Quick spin the cellls with primary antibody. Place the cells with primary antibody on a magnetic stand, carefully remove the solution.
Add 1ml Dig-Wash Buffer. Invert the tube 10 times to resuspend the beads.
Repeat step 20-21 twice.
Bind pA-M.EcoGII recombinant enzyme
Add 20ul pA-M.EcoGII recombinant enzyme into 130ul dig-wash Buffer, mix gently by pipetting.
Quick spin the tube from step 22. Place the tube on a magnetic stand, carefully remove the solution.
Add the pA-M.EcoGII containing buffer to the cells with gentle vortexing.
Place the tube on end-over-end rotator at room temperature for 1h.
Quick spin the cells with pA-M.EcoGII. Place the cells with pA-M.EcoGII on a magnetic stand, carefully remove the solution.
Add 1ml Dig-Wash Buffer. Invert the tube 10 times to resuspend the beads.
Repeat step 27-28 twice.
Methyltransferase activation
Quick spin the tube from step 29. Place the tube on a magnetic stand, carefully remove the solution.
Add 300ul Methylation Buffer. Invert the tube 10 times to resuspend the beads.
Incubate at 37°C thermomixer at 300rpm for 30min. Supplement 7.5ul 32mM SAM at 7.5min, 15min, and 22.5min.
DNA extraction:PCI
Take the tube from step 32. To stop the methylation reaction, add 10ul 0.5M EDTA, 1.5ul 20% SDS, and 5.0ul 20mg/ml Proteinase K to each tube.
Mix by vortexing at highest speed for 5s. Incubate at 55°C water batch overnight until the solution is clear.
Note: Increase incubation time if the solution is viscous or cloudy.
Add 300ul PCI and vortexing at highest speed for 30s. Invert the tube 10 times to mix thoroughly. Centrifuge at 16000g for 5min.
Transfer the upper liquid aqueous phase to a new 1.5ml centrifuge tube. Add 300ul chloroform. Invert the tube 10 times. Centrifuge at 16000g for 5min.
Transfer the upper liquid aqueous phase to a new 1.5ml centrifuge tube. Add 1/10 volume of 3M sodium acetate solution.
Add 2.5x volume of ice cold 100% ethanol in the solution to precipitate DNA. Incubate the tube at -20 overnight.
Centrifurge at 16000g for 10min. Discard the supernatant and rinse the pellet with 70% cold ethanol.
Air-dry the pellet. Dissolve in TE buffer.
DNA extraction: Serapure Beads
Take the tube from step 32, place the tube on a magnetic stand, remove all the solution.
Add 600ul pre-warmed Digestion Buffer, plus 4.0ul 10mg/ml RNase A. Mix thoroughly immediately by pipetting up and down 10 times with a wide-bore tip.
Add 10.0ul 20mg/ml Proteinase K. Incubate the tube at 55°C water bath overnight.
Add 200μl (or 1/3 of the lysis buffer volume) of 5M potassium acetate and mix by inverting the tube 20 times in order to obtain a homogenous solution to fully precipitate the proteins and the polysaccharides that will complex with SDS. It is important to incubate at 4°C after the addition of potassium acetate.
Centrifuge at 5000g for 10 minutes at 4°C. Transfer the supernatant to a new 1.5 ml tube without disturbing the pellet.
Add one volume of Bead Binding Buffer and 1:18 (v:v) of Serapure beads previously prepared (vortex the beads solution for 20 seconds before use to ensure that the beads are completely resuspended).
Mix by inverting the tube 20 times. Incubate with a gentle mixer for 10 minutes at room temperature.
Quick spin the tube to remove the liquid from the cap. Place the tube in a magnetic stand until the solution becomes clear (2-3min). The actual time required to collect beads may vary according to samples.
Remove the supernatant without disturbing the beads pellet. Add 1 ml of Bead Wash Buffer, remove the tube from the magnetic rack and mix by inverting the tube 20 times.
Quick spin the tube to remove the liquid from the cap. Place the tube in a magnetic stand until the solution becomes clear (2-3min).
Repeat step 49-50.
Quick spin the tube to remove the liquid from the cap. Place the tube in a magnetic stand to remove the remaining Bead Wash Buffer.
Let the beads air-dry for 1 minute with the cap open. Do not let the beads dry more than 1 minute as this will significantly decrease elution efficiency.
Add 80 μl of TE buffer preheated to 50°C.
Resuspend the beads by flicking the tube. It is important that the beads are not aggregated.
Quick spin the tube. Place the tube in the magnetic rack. Let the solution to become clear. If DNA solution is highly concentrated, it can take a long time. In this case, it is recommended to let the tube in the magnetic rack overnight or to add more elution buffer.
Transfer 75 μl of the eluted gDNA solution in a new tube.
ONT library prepartion and sequencing
Follow Oxford Nanopore protocol of LSK109 for adaptor ligation.
Load 500ng of the ligated DNA to R9.4.1 flow cell. Use the Flow Cell Wash Kit to wash the flow cell.
Reload the flow cell every 24h.

