Automated Protein Normalization and Tryptic Digestion on a Biomek-NX Liquid Handler System
Yan Chen, Nurgul Kaplan Lease, Jennifer Gin, Tad Ogorzalek, Christopher J Petzold
Abstract
This protocol details steps to normalize the amount of protein for tryptic digestion in quantitative proteomic workflows by using a Biomek NX liquid handler system. It is optimized to normalize protein concentrations in a 96-well plate format and add TCEP, IAA, and trypsin.
This protocol works best as part of a semi-automated proteomic sample preparation workflow with:
Automated Chloroform-Methanol Protein Extraction on the Biomek-FX Liquid Handler System
and
Automated Protein Quantitation with the Biomek-FX liquid handler system
Before start
For this protocol you will need:
-
A Beckman-Coulter Biomek NX-S8 or NXP liquid handler system with a 8-pod head
-
Upload the attached method files and modify them to fit your deck and system configuration.
Proteomics-Normalization Method.bmf Proteomics-Plate to Plate Transfer.bmf
- Protein samples of known concentration
Chemicals to prepare:
-
Prepare
100millimolar (mM)by dissolving28.7mgin1mL -
Prepare
200millimolar (mM)by dissolving36.8mgin1mL -
Prepare
1mg/mLby adding1mLto1mgthen vortex to mix
Steps
Biomek NX-S8 input file preparation
After measuring protein concentration by the DC (Detergent Compatible) protein assay (Bio-Rad), export protein concentration report through MD Spectramax 250 software that controls the microplate reader. Copy the content in the exported text file and paste it into Excel, and then save as a UTF-8 format text file.
Use MS Excel or a Jupyter notebook to normalize the protein concentration to 50µg and convert the spectrophotometer output file into the following two files in a format suitable for the Biomek NX-S8:
NX-AMBIC.csv
| A | B | C | D | E |
|---|---|---|---|---|
| scrpos | srcwell | destpos | destwell | vol |
| media | 1 | DestPlate | 1 | 33.05 |
| media | 1 | DestPlate | 2 | 26.73 |
| media | 1 | DestPlate | 3 | 27.96 |
| media | 1 | DestPlate | 4 | 28.74 |
| media | 1 | DestPlate | 5 | 22.94 |
| media | 1 | DestPlate | 6 | 28.07 |
| media | 1 | DestPlate | 7 | 24.34 |
| media | 1 | DestPlate | 8 | 28.12 |
| media | 1 | DestPlate | 9 | 26.64 |
| media | 1 | DestPlate | 10 | 26.22 |
NX-AMBIC.csv output table
NX-protein.csv
| A | B | C | D | E |
|---|---|---|---|---|
| scrpos | srcwell | destpos | destwell | vol |
| SrcPlate | 1 | DestPlate | 1 | 10.95 |
| SrcPlate | 2 | DestPlate | 2 | 17.27 |
| SrcPlate | 3 | DestPlate | 3 | 16.04 |
| SrcPlate | 4 | DestPlate | 4 | 15.26 |
| SrcPlate | 5 | DestPlate | 5 | 21.06 |
| SrcPlate | 6 | DestPlate | 6 | 15.93 |
| SrcPlate | 7 | DestPlate | 7 | 19.66 |
| SrcPlate | 8 | DestPlate | 8 | 15.88 |
| SrcPlate | 9 | DestPlate | 9 | 17.36 |
| SrcPlate | 10 | DestPlate | 10 | 17.78 |
NX-protein.csv output table
Biomek NX-S8 preparation
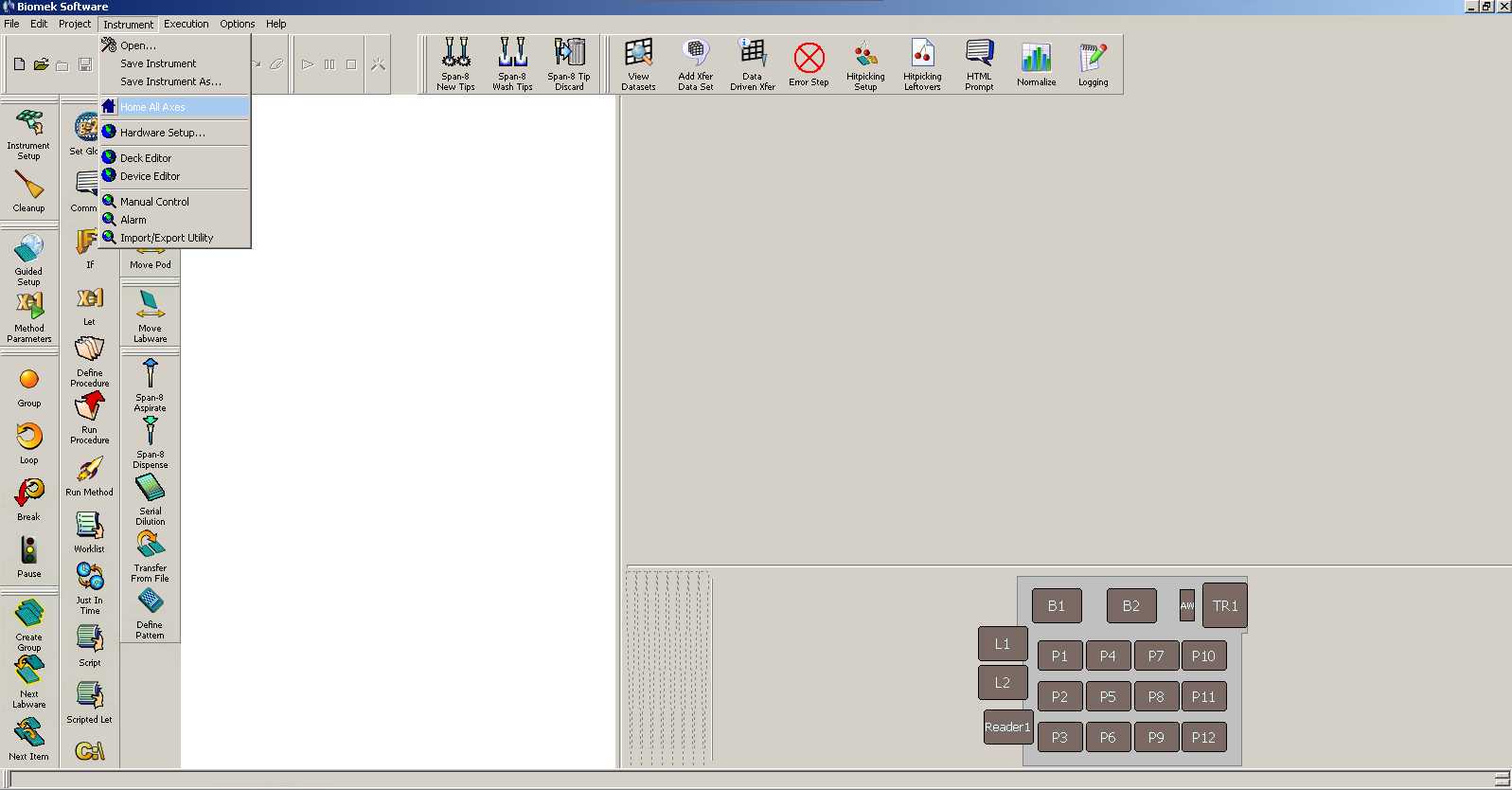
Open Biomek software program from Biomek NX-S8 control computer. In the "Instrument" drop-down menu, select "Home all Axes" to prepare the instrument for use and purge air from the tubing and syringes.


Buffer Transfer
Go to "Open Method." Select the "Proteomics" folder and open the method "Proteomics-Normalization Method."
Set up the deck (refer to the deck setup picture above):
| A | B | C |
|---|---|---|
| Deck Label | Labware | Reagent |
| DestPlate | PCR plate 96-well non-skirted (Thermo Fisher, Cat.#AB0600) on a yellow PCR rack | |
| AMBIC | Biomek Reservoir (discontinued) | Ammonium Bicarbonate buffer |
| tips | 200 uL pipet tips (Molecular Bioproducts BioRobotix, Cat.#919-262 ) |
Deck materials
Click on "Transfer From File."
Copy the NX-AMBIC.csv file generated in Excel or via a Jupyter Notebook into the directory that the method is designated to read. For example *C:\Users\jbei\Desktop\Proteomics Methods\CSV files*
Click on the 2nd "View Datasets" to check that you have copy and pasted the correct volumes in the 96-well format.
Click "Finish" to make sure there are no error messages.
Click the "Run" button (green arrow) to start.
Protein Transfer
Go to "Open Method." Select the "Proteomics" folder and open the method "Proteomics-Plate to Plate Transfer."
Set up the deck (refer to the deck setup picture above):
| A | B | C |
|---|---|---|
| Deck Label | Labware | Reagent |
| DestPlate | PCR plate 96-well non-skirted (Thermo Fisher, Cat.#AB0600) on a yellow PCR rack | Ammonium bicarbonate buffer |
| SrcPlate | PCR plate 96-well non-skirted (Thermo Fisher, Cat.#AB0600) on a yellow PCR rack | protein |
| tips | 20 uL pipet tips (Molecular Bioproducts BioRobotix, Cat.#918-262 ) or | |
| 200 uL pipet tips (Molecular Bioproducts BioRobotix, Cat.#919-262 ) |
Deck materials
Click on "Transfer From File."
Copy the NX-protein.csv file generated by Excel or via a Jupyter Notebook into the directory that the method is designated to read. For example: *C:\Users\jbei\Desktop\Proteomics Methods\CSV files*
Click on "View Datasets" to check that you have copy and pasted the correct volumes in the 96-well format.
Click "Finish" to make sure there are no error messages.
MANUAL STEP: Use a multichannel pipette to mix protein samples completely right before starting.
Click the Run button (green arrow) to start.
Trypsin Digestion
Chemicals to prepare:
-
Prepare
100millimolar (mM)by dissolving28.7mgin1mL -
Prepare
200millimolar (mM)by dissolving36.8mgin1mL -
Prepare
1mg/mLby adding1mLto1mgthen vortex to mix
Add the following reagents to the normalized protein plate, in this order:
2.5µL2.5µL1µL
Incubate at 37°C for 4h 0m 0s to 16h 0m 0s.
Clarifying Digestion Reaction
After digestion, spin at 14000rpm .
Pipet supernatant into new PCR plate. Seal and store at -20°C until ready for LC-MS/MS analysis (e.g., Discovery Proteomics protocol, Targeted Proteomics protocol).