A versatile nuclei extraction protocol for single cell multiome ATAC and gene expression in non model species
Rose Ruiz Daniels, Richard S Taylor, Sarah Salisbury, Emily Clark, Dan Macqueen, Diego Robledo, Ioannis Konstantinidis, Diego Perojil Morata, Jorge Manuel de Oliveira Fernandes
Disclaimer
DISCLAIMER – FOR INFORMATIONAL PURPOSES ONLY; USE AT YOUR OWN RISK
The protocol content here is for informational purposes only and does not constitute legal, medical, clinical, or safety advice, or otherwise; content added to protocols.io is not peer reviewed and may not have undergone a formal approval of any kind. Information presented in this protocol should not substitute for independent professional judgment, advice, diagnosis, or treatment. Any action you take or refrain from taking using or relying upon the information presented here is strictly at your own risk. You agree that neither the Company nor any of the authors, contributors, administrators, or anyone else associated with protocols.io, can be held responsible for your use of the information contained in or linked to this protocol or any of our Sites/Apps and Services.
Abstract
Here we present a modified version of : dx.doi.org/10.17504/protocols.io.261genwm7g47/v2 that was used to successfully extract nuclei from an array of different tissue types for single cell sequencing and modify it with the purpose of extracting nuclei for single cell multiome ATAC and gene expression on the 10x chromium. The modifications for this protocol include: different concentration of RNase inhibitors, different quantities for nuclear isolation buffer, removal of unnecessary steps as well as QC specific for multiome analysis.
If you are looking to use this protocol for bulk ATAC-seq use of protease inhibitor cocktail PIC is recommended instead of RNase inhibitor on the snRNA-seq version of this protocol (dx.doi.org/10.17504/protocols.io.261genwm7g47/v2) please get in touch with the authors if you are unsure on how to do this.
Before start
Sampling and storage for nuclear isolation.
Animals must be appropriately euthanized and immediately processed. Approximately ~60mg of tissue is placed in one clearly labelled cryotube and immediately flash frozen in liquid nitrogen. This step is critical . The tissue must be preserved as fast as possible for optimal results. In the absence of liquid nitrogen, samples can be frozen in dry ice. Samples can be stored at -80°C for up to a year prior to use. Older samples might still yield viable nuclei but this would need to be tested.
All reagents should be chilled on ice prior to use.
Samples should be kept frozen on dry ice until immediately before nuclei isolation, and all sample-handling steps should be performed on ice.
The centrifuge should be pre chilled at 4°C.
All reagents are given for 2 nuclear isolations.
Amounts of buffer especially those that contain RNase should be adjusted appropriately for each experiment prepared prior and RNase added immediately before use.
Steps
Nucleus isolation workflow for ST-based buffers
On ice, place a piece of frozen tissue into one well of a 6-well tissue culture plate with 1mL TST.
On ice, mince tissue initially using Tungsten Carbide scissors for 0h 0m 30s and then with Noyes Spring Scissors 0h 10m 0s.
0h 5m 0s into the mincing gently pipette up and down with a p1000 pipette using a low retention filtered tip. The time in the dissociation buffer is critical. See image for how to assess the timing is correct by looking at your nuclei.
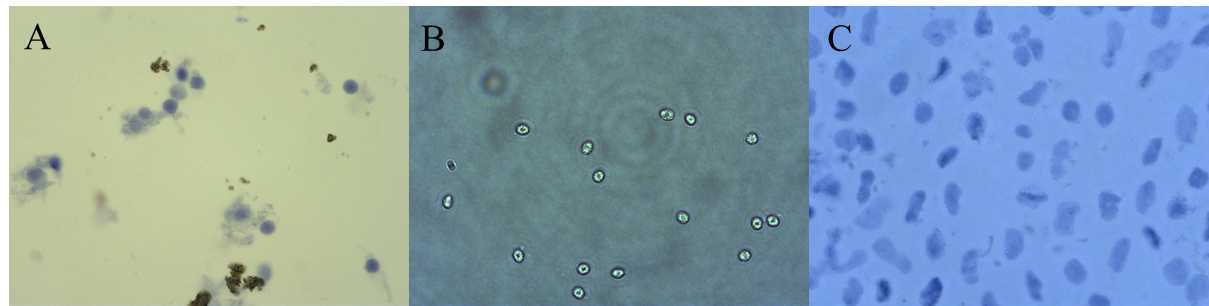
Pass lysate through a 40 µm cell strainer .
Add a further 1mL of TST to the cell strainer immediately.
Add 3mL of freshly prepared ST buffer to the lysate.
Add the 5mL of lysate to a marked 15 ml falcon tube (Corning) on ice.
Centrifuge at 500x g,4°C in a swinging bucket centrifuge.
Discard liquid, carefully remove excess liquid with a p200 pipette, careful to not disturve the pellet. Resuspend the pellet gently using a p1000 pipette in diluted nuclei buffer in aiming for target recover of 6.000 nuclei. The concentration of RNAse inhibitor should be 1U/ul.
Count the nuclei using a C-chip disposable haemocytometer.
The nuclei are also counted using a Bio-Rad TC20 to confirm results from the disposable haemocytometer and to count the proportion of viable cells.

