A simple illustrated protocol to prepare over 1,000 multiplexed single cell proteomics samples per day
Benjamin Orsburn
Disclaimer
This is for research purposes only.
TMT is a registered trademark of Proteome Sciences LLC
No guarantees are stated or implied.
Abstract
This is a simple illustrated sample preparation method for multiplexed single human cells, as described in Orsburn et al., Bumpus 2022 that better encompasses the lab SOP I have developed over the last couple of years.
With no specialized equipment or robotics, I can easily, as a single operator prepare, lyse, digest and label over 1,000 single human cells per day with just the help of a sorting core to place single human cells into plates as indicated in this tutorial.
If I try to do more than 2,000 single cells in a day, chances are I'm going to mess up more than a few plates.
I have provided this protocol to students with no prior experience in the preparation of proteomics samples and they have successfully prepped multiple plates per day.
This protocol should be amenable to rapid and inexpensive automation.
A full tutorial on this protocol is in works, look out for it in 2023 and please contact me if you have questions or concerns.
This protocol is a brazen adaptation of Budnik et al., 2018 in the simplest possible form to make SCOPE-MS approachable to even scientists with no experience in proteomics sample prep.
Steps
Prepare 96-well plates for sorting with acetonitrile lysis solution
Add 2uL of LCMS grade acetonitrile to each well of a 96 well plate
Centrifuge if necessary to ensure acetonitrile is in the bottom of the plate. 1 minute pulsing is more than sufficient. A salad spinner based centrifuge can complete 4 plates/minute.
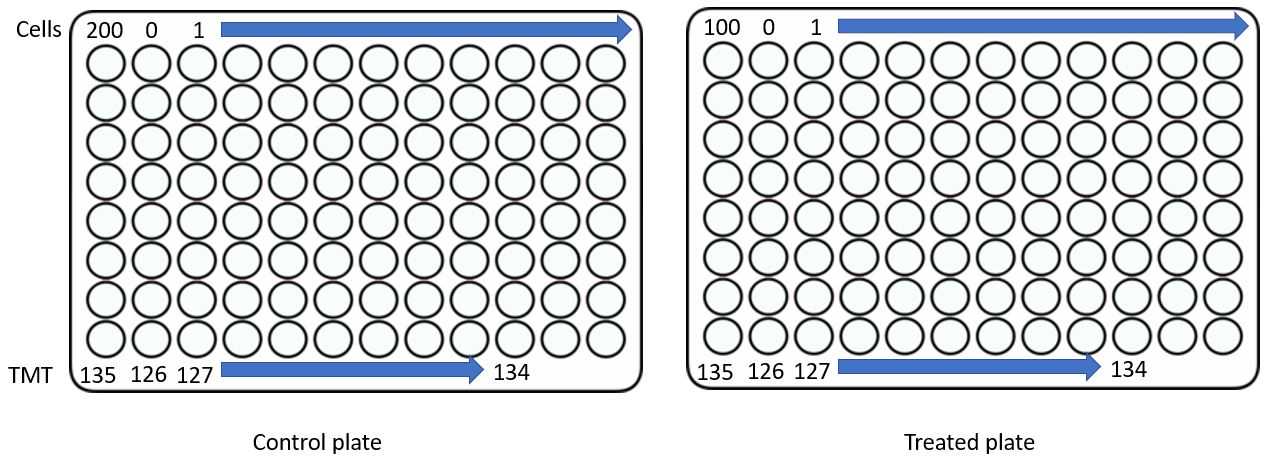
Using flow cytometry or microscopy based sorting, aliquot cells into plates using the following template. In this case we have two conditions, so control cells and drug treated cells are separated. These will be combined in a later step.
IMPORTANT NOTE: This experiment was designed for pasefRIQ on a TIMSTOF with a generation 2 TIMS cartridge. I have observed no "carrier proteome effects" when using 200 cell loads on this device. The number of cells may need to be adjusted down for instruments with more pronounced carrier proteome limits, in particular D20 Orbitrap systems.
Once cells have been aliquotted into plates. Seal these tightly and place the plates directly onto dry ice for at least 30 seconds. The frozen and lysed cells may then be stored on wet ice until moved to -80C storage.
Prepare reagents
First prepare the digestion buffer by adding 50 uL of 1M TEAB + 10mg of DDM (n-Dodecyl-beta-Maltoside Detergent, Thermo Fisher 89902) in 10mL of LCMS grade water.
Chill this solution on wet ice.
To prepare trypsin at 5ng/uL add 4mL of digestion buffer to 20 micrograms of LCMS grade trypsin (we use Pierce 90057, but there are many alternatives). Storage of this solution on wet ice reduces self-digestion of the enzyme, but likely is not critical.
Preparing cells for digestion
Prepare a heat block of thermocycler at 95C for drying off acetonitrile and further disrupting the cell membranes.
Remove cells from -80C storage and immediately place these on the heat block, removing the air tight seal as you do so. 1 minute will be enough to drive off the acetonitrile and to obtain an internal well temperature of 95C in most conditions.
Add trypsin solution
*Using a multichannel pipette is the easiest here, but not essential for low cell numbers or optimization experiments
I pour the trypsin solution into a sterile multichannel trough (Heaththrow Scientific 120637) and pipette 1 microliter of this solution into each well for a total of 5ng of trypsin per well.
In this protocol I'm using trypsin as the "sacrifical carrier" (please see Sam Choi, 20222 for details on this concept).
Seal the plate securely. In my hands, silicone web seals do not hold in condensation successfully for low volumes. Adhesive plate seals are preferable (Fisher 60180-M143, for example. A plate seal roller may be helpful, but not necessary.
Incubate for digestion
Multiple methods work well here.
45C for 2 hours with gentle shaking is very effective (Orsburn et al., 2022) particularly if the plates are centrifuged every hour to keep condensation down.
Room temperature overnight can also be effective. However, if the plates are not going to be attended, you may want to increase the total digestion volume by adding 2 microliters of the digestion buffer.
**I do not recommend high temperature overnight. The samples will simply evaporate and the reaction may not occur to completeness.
Dry plates if proceeding to label free analysis, move to step 13 if TMT labeling
Following incubation, centrifuge plates to concentrate all liquid.
If TMT (SCoPE-MS, pasefSCOPE, iBASIL) Move to Step 13.
Dry off the digestion buffer with vacuum centrifugation.
*If necessary, I have found when my speedvac was broken and Eppendorf could not repair it for extended periods of time, the volume may be driven off at 55C on a heat block.
**If you are doing a label free analysis you are done. Load 2 microliters of 0.2% formic acid into each well, vortex, centrifuge and load directly.
If you are labeling proceed to 13.
Prepare TMT reagents at appropriate concentrations
A single TMT kit with 0.5mg of reagent per vial (such at Thermo A44522) can label more than 60,000 single cells. If you are exclusively doing single cell prep, I suggest making multiple aliquots once opening a kit.
Add 120 uL of LCMS grade acetonitrile to each 0.5mg vial of TMT reagent to be used.
If aliquoting, transfer 10 uL of each to sterile microcentrifuge tubes and rapidly dry via speedvac.
For single cell labeling concentrations, dilute each 10uL aliquote by adding 190 microliters of LCMS grade acetonitrile. This creates a 20ng/uL solution.
Label cells with TMT
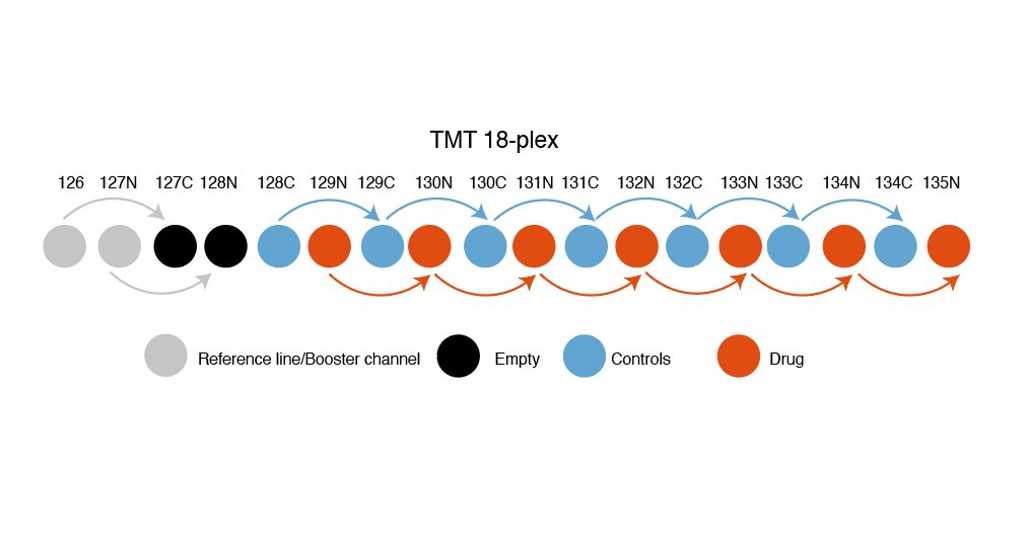
Add 4 microliters of the TMT label to your carrier channel if 200 cells were used. This will saturate your peptides with approximately 4:1 TMT reagent
Add 1 microliter of the TMT label to each of your single cell digests and blank well. This will provide 4:1 for the trypsin peptides that are present, as they are in significantly higher concentration than your single cell peptides, this step is critical.
Label for 20 minutes at room temperature with gentle shaking.
Centrifuge all solution to concentrate solution.
Quench each reaction by adding 2 uL of 0.5% hydroxylamine to your carrier channel and 0.5 uL of 0.5% hydroxylamine to each single cell and blank well. 15 minute reaction at room temperature is sufficient.
Wells may be combined now or stored at -80C until combining.
*I have found single cell peptides may not be very good after a few weeks of storage. This is not quantitative, simply an observation.
Combining cells for multiconditional experiments
If you are not doing multiconditional experiments you can simply combine your wells across.
If you are doing two conditions, follow these steps
First create a combined carrier channel that is an equal mixture of your digested and labeled control and treatment conditions.
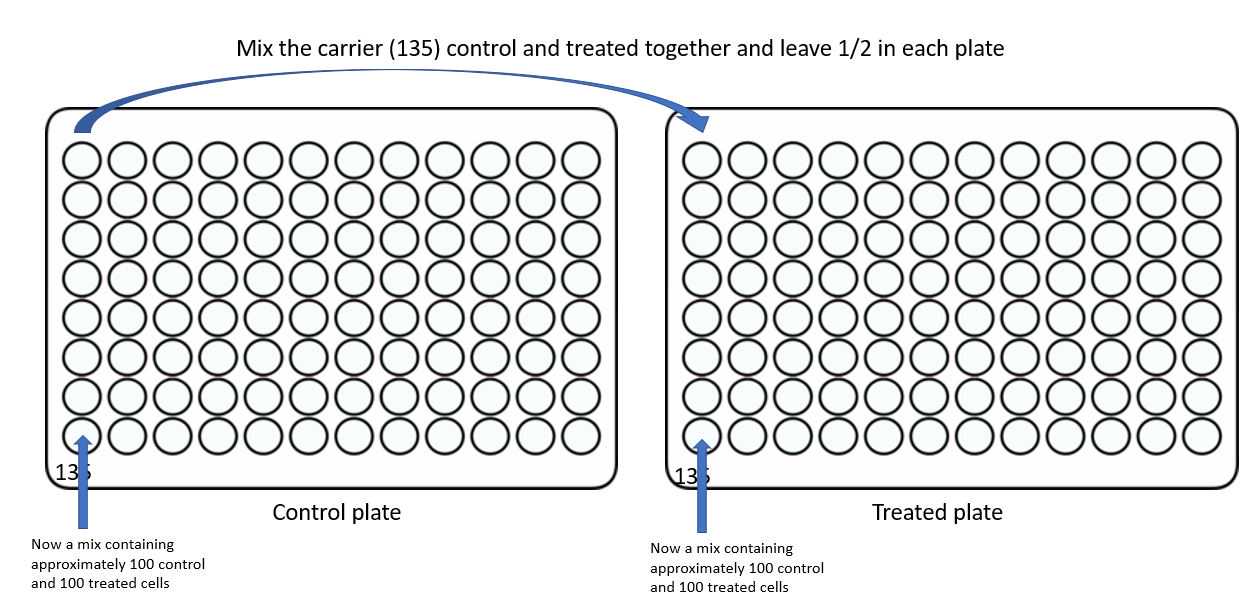
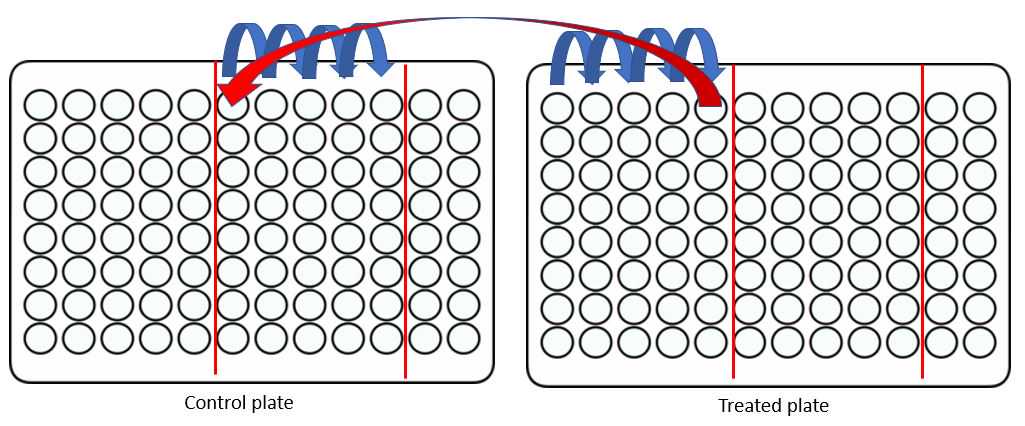
Now combine 5 wells from the control plate with 5 wells from the treated plate:
*Note, I find it helpful with my poor eyesight and bifocals to mark each plate with a sharpie as shown.
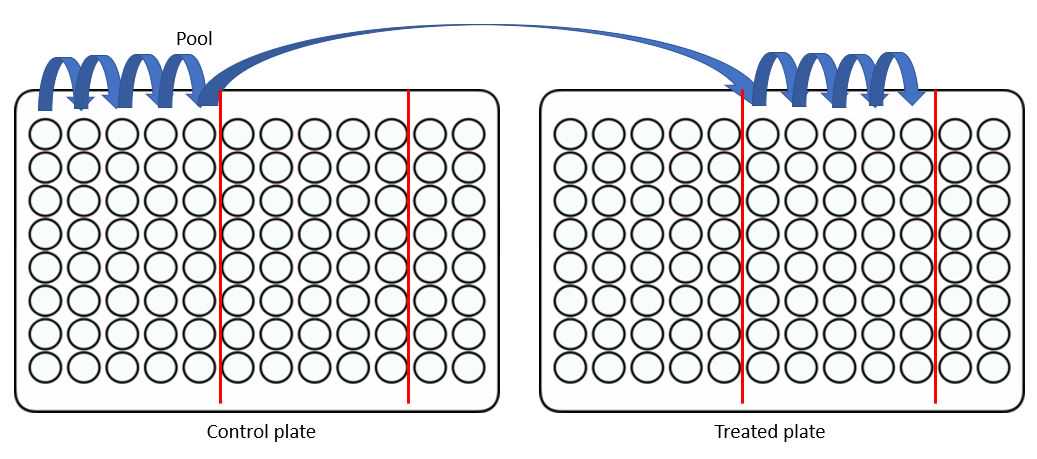
The final (10th well) of each plate can now be transferred to a new plate for drying and sample injection. I find it preferable to stack my samples alternating, so that, in this example A1-H1 will be the samples prepared in step 27 and A2-H2 are the samples prepared in 28. Running samples A1 then A2 will have alternating patterns of control and treated cells which are great for improving your confident that results are not batch related effects.
To get to approximately 1,000 single cells prepared/day you will need to prepare 16 plates at a time. On my own I have found more than 24 to be difficult to manage, but I'm also quite old and partially blind in one eye.