A scalable hydroponic-based method for screening isolates of chickpea Fusarium wilt pathogen, Fusarium oxysporum f.sp. ciceris
Manavi Raizada, Nimmy MS, Ramawatar Nagar
Abstract
Chickpea is the third most consumed grain legume in the world. It serves as a valuable source of protein and micronutrients, especially for the large vegetarian population in the Indian subcontinent. One of the major constraints in chickpea production is the Fusarium wilt disease caused by the soil-born necrotrophic pathogen Fusarium oxysporum f. sp. ciceris (Foc). The pathogen has been shown to interact with chickpea in a race-specific manner and evolved into multiple races. So far eight races of it have been reported including 0, 1A, 1B/C, 2, 3, 4, 5, and 6. When a pathogenic race and host genotype interact in a race-specific manner, the interaction follows the typical gene-for-gene interaction (R-AVR) hypothesis, where major genes control avirulence/virulence in the pathogen and resistance/susceptibility in the host. However, so far neither any race-specific molecules (AVR) from the Fusarium oxysporum f. sp. ciceris nor the resistance gene (R) from the chickpea have been reported.
We are using a forward genetics screen for the identification of avirulence genes by r and t-DNA insertional mutagenesis.
Since Fusarium is a soil-born pathogen that infects roots, the usual root dip method used for its screening cannot scale for screening 1000 mutants. Hence, the need arises for a scalable high throughput method to screen numerous mutants.
We are in the process of optimizing a hydroponics-based screening method of pathogens. We have tried to optimize many conditions for a highly reproducible method.
Steps
Seed Germination
An approximately equal number of seeds of both resistance and susceptible genotypes, JG-62 and WR-315, were taken in 2 separate flasks. They were washed first with 0.042 % (w/v) sodium hypochlorite solution (42 µl sodium hypochlorite in 100 ml solution with double distilled water) and then 4–5 times with double distilled water to remove the leftover chemical.
In wide pre-sterilized Petri plates layered with autoclaved blotting paper, 1 plate for each genotype, the sterilized seeds were spread out and a little autoclaved double distilled water was added. These plates were kept in a culture room covered with foil and the seeds were allowed to germinate in the dark for about 5–6 days. Double distilled water was added from time to time to ensure optimum water levels and seed germination.

Hydroponics System (HS) preparation Hydroponics System (HS) preparation -
All glassware was thoroughly washed with Cedepol cleaning agent, dried in a hot air oven, and then autoclaved before use. The foam sheet was sterilized in diluted Cedepol cleaning agent by soaking for 15–20 minutes and then rinsing with distilled water until the water runs clean.
Plugs the size of the mouth of the test tube were cut out. Using a lamp filled with 70 % ethanol and a pair of forceps heated at the ends, twin holes were poked into each plug.

Transfer to Hydroponics system
After the seeds had germinated, they were transferred to the hydroponics system. 2 seeds of a genotype were placed into the twin holes of 1 foam plug using forceps, which was then fit into a long wide-mouthed test tube filled with ½ strength Hoagland’s No. 2 Basal salt solution (Figure 3).
Triplicates were set up for each isolate (that would be used for infection later) plus a control tube. This was done for both genotypes (Figure 3).

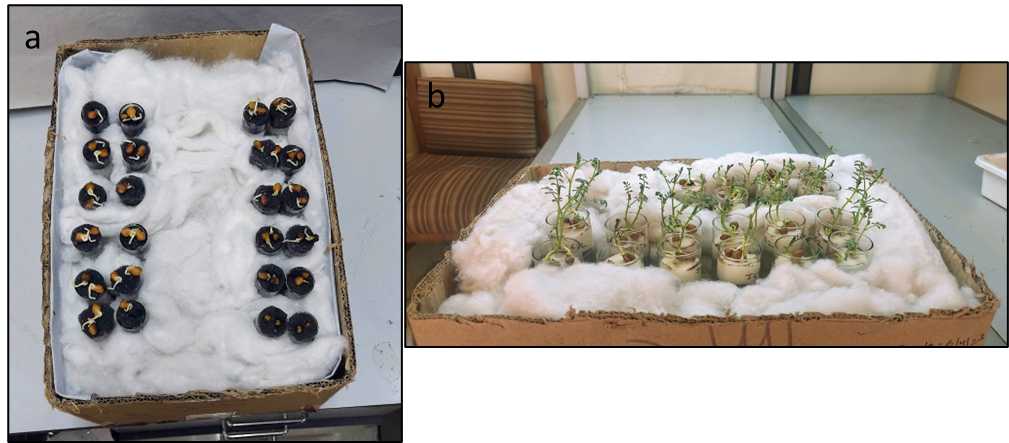
All tubes were correctly labeled with the genotype they were holding with the help of tough tags. These tubes held in a stand were placed in a cardboard box stuffed with cotton to ensure a dark environment below the plugs (similar to a soil system) (Figure 4(a)).
After shifting to the HS system, the seedlings were allowed to grow, their water levels maintained, and were then infected when they were 10–12 days old since germination (Figure 4(b)).
Infection with FOC isolates :
The FOC isolates, previously inoculated, filtered, and the spores counted (Figure 5), were diluted with double distilled water (if required) to reach a concentration of 106 – 108 spores /ml.

This spore solution was dispensed into the tubes with a micropipette, triplicates for each isolate for both genotypes. Nothing was added to the control tubes. They were correctly labeled on the tough tags.
These infected seedlings were observed for symptoms of infection and wilting, and optimal water levels and growth conditions were maintained throughout.
Differential Phenotypes
With the results obtained so far, it can be generalised that JG62 showed stunted growth with less dense foliage and roots, contrary to WR315 that had thick growth (Figure 6). Their respective control tubes displayed equally optimal growth irrespective of their genotypes.


