ASAP protocol: Crystallization of Enterovirus D68 3C protease (PDB code 8CNX)
Peter Marples, Lizbé Koekemoer, Daren Fearon, ryan Lithgo
Abstract
Steps
Equipment needed
Formulatrix Rock Imager (or incubator of choice)
P100 multi-channel pipette
Crystallization experiment
Protein and buffer requirements:
32µL``1millimolar (mM) Sample
2.88mL
16µL Sample seeds, dilution 1:100 000
Crystallisation screen composition:
0.1 M Tris-HCl (pH 8.14)
0.2 M Ammonium acetate
25% w/v PEG 3350
Stock solutions used:
1 M Tris-HCl adjusted to pH 8.14 with HCl
1 M Ammonium acetate
50% w/v PEG 3350
Prepare seed stock: seed stock:
1: 100 000 dilution Sample seeds
Dispense 30µL into SwissCI 3 lens plate reservoir wells using a 100 µl multi-channel pipette.
Dispense 100nL``1millimolar (mM) Sample to each lens using the SPT mosquito.
Dispense 200nL to each lens using the SPT mosquito.
Dispense 50nL to each lens using the SPT mosquito.
Drop ratio: 1:2:05 ratio (100 nl Sample : 200 nl reservoir solution: 50 nl seeds)
Final drop volume: 350 nl
Incubate at 20°C for 24h 0m 0s in Formulatrix Rock Imager.
Imaging Schedule : The first images are taken after 12 h and the imaging schedule follows a Fibonacci sequence of days for further collections.
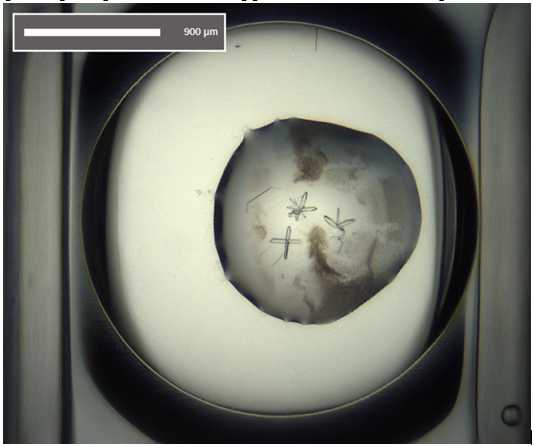
Crystal form after ~12 h.