814.1 Multiplexed Immunofluorescence Phenocycler-Fusion® Imaging of FFPE Lung Sections Following Mass Spectrometry
Gloria S Pryhuber, Heidie Huyck, Jeffrey Purkerson, Luis Colon
Abstract
This protocol describes multiplexed immunofluorescent (MxIF) staining and imaging of FFPE lung tissue sections utilizing the Phenocycler-Fusion platform (Akoya Biosciences) following spatial analysis of N-Glycans with MALDI-MSI. The approach for MxIF is based on the CODEX multiplexed immunofluorescence staining technology developed by Garry Nolan and colleagues (1). The protocol is closely aligned with the attached Phenocycler-Fusion User Guide provided by Akoya (2).
The protocol includes A) FFPE lung tissue section preparation, B) Reference to spatial N-glycomics with MALDI-MSI for human lung tissue C) Labeling of lung tissue sections with antibody-barcode conjugates, D) Barcode-Reporter plate and experiment design, E) Multiplexed imaging and analysis, F) Reference for H&E stain to follow MxIF and G) Reference to custom antibody conjugation. Thus, the protocol is designed to provide information regarding specific reagents (i.e., antibodies), conditions (i.e., dilutions), and procedures used in multiplexed immunofluorescent staining of lung tissue.
Before start
The Phenocycler Fusion system must be setup and calibrated by an installation engineer. Akoya Biosciences will provide a list of materials required for training.
Attachments
Steps
Lung Processing and Sectioning
FFPE Lung sections (5-7 µm) are prepared as described in dx.doi.org/10.17504/protocols.io.kxygxejwdv8j/v2 and mounted on indium-tin-oxide (ITO) glass slides (25 mm x 75 mm) from Delta technologies (http://www.delta-technologies.com/Products.asp?c=13&s=17) Part #: CG-80IN-S115.
Spatial N-glycomics with MALDI-MSI for human lung tissue
N-linked Glycans are detected in FFPE lung sections via MALDI-Mass Spectrometry as described in dx.doi.org/10.17504/protocols.io.5jyl8p876g2w/v2.
Post MALDI Imaging MS acquisition, MALDI matrix is removed by incubation in 2X Coplin Jars containing 50% acetonitrile.
- 50% Acetonitrile #1
0h 2m 0s - 50%Acetonitrile #2
0h 2m 0s
Slides were air-dried and desiccated prior to shipping back to URMC.
Labeling of lung tissue sections with antibody-barcode conjugates
The tissue sections are rehydrated through descending ethanol series followed by molecular biology grade distilled water (2X).
- 100% Ethanol #1
0h 5m 0s - 100% Ethanol #2
0h 5m 0s - 90% Ethanol
0h 5m 0s - 70% Ethanol
0h 5m 0s - 50% Ethanol
0h 5m 0s - 30% Ethanol
0h 5m 0s - ddH20 #1
0h 5m 0s - ddH20 #1
0h 5m 0s
High pH Antigen Retrieval 9
Heat-Induced Epitope Retrieval (HIER) reverses protein crosslinking in FFPE tissue
Dilute AR9 buffer (Akoya Biosciences) 1/10 in ddH20 and fill plastic Coplin Jar to 90-95% volume. Add slides containing tissue sections to be stained. Cover entire Coplin Jar with aluminum foil. (Do not Cap)
Place Coplin Jar in an InstantPot® pressure cooker containing ddH20 to 1/3-1/2 depth of Coplin Jar.
Heat on high pressure setting 0h 20m 0s
After releasing pressure per pressure cooker directions, remove Coplin Jar, partially unwrap foil without uncovering slides and allow slides to cool for a minimum of 1h 0m 0s . Attempting to rinse/wash without allowing slides to cool may reduce tissue adherence to glass slide.
Prepare Blocking Buffer
Prepare Blocking Buffer no earlier than 1 h before staining (i.e. while slides are cooling in AR9 buffer and/or incubating in Staining buffer, see below) and keep on ice.
Table 1. Blocking Buffer Component Table
Component 2 Slides 5 slides N Slides Staining Buffer 362µL 905µL 181µL N Blocker 9.5µL 23.75µL 4.75µL G Blocker 9.5µL 23.75µL 4.75µL J Blocker 9.5µL 23.75µL 4.75µL S Blocker 9.5µL 23.75µL 4.75µL Total Volume 400µL 1000µL 200µL
Prepare Antibody Cocktail
In a 1.5 ml microfuge tube, mix volume of blocking buffer = 200µL minus total volume of antibodies (µl/slide x N) to be used in experiment (See Table below). The Blocking buffer volume must be ≥ 60% of total antibody cocktail volume for effective blocking. If needed, reduce staining buffer volume to achieve 60% blocking buffer volume in Ab solution.
Example antibody panel (final column = microliter of antibody per single slide):| A | B | C | D | E | F | | --- | --- | --- | --- | --- | --- | | SMA | Akoya | 4450049 | BX013 | 1:200 | 1 | | PanCK | Akoya | 4450020 | BX019 | 1:200 | 1 | | MPO | Akoya | 4250083 | BX098 | 1:200 | 1 | | Ki67 | Akoya | 4250019 | BX047 | 1:200 | 1 | | Keratin5 | Akoya | 4450090 | BX101 | 1:200 | 1 | | HLADR | Akoya | 4550118 | BX033 | 1:200 | 1 | | FOXP3 | Akoya | 4550071 | BX031 | 1:200 | 1 | | ColIV | Akoya | 4550122 | BX042 | 1:200 | 1 | | CD8 | Akoya | 4250012 | BX026 | 1:200 | 1 | | CD68 | Akoya | 4550113 | BX015 | 1:200 | 1 | | CD45 | Akoya | 4550121 | BX021 | 1:200 | 1 | | CD4 | Akoya | 4550112 | BX003 | 1:200 | 1 | | CD3e | Akoya | 4550119 | BX045 | 1:200 | 1 | | CD31 | Akoya | 4450017 | BX001 | 1:200 | 1 | | CD20 | Akoya | 4450018 | BX007 | 1:200 | 1 | | CD163 | Akoya | 4250079 | BX069 | 1:200 | 1 | | CD14 | Akoya | 4450047 | BX037 | 1:200 | 1 | | CD11c | Akoya | 4550114 | BX024 | 1:200 | 1 | | E-Cadherin | Akoya | 4250021 | BX014 | 1:200 | 1 | | TPSAB1* | Abcam | ab2378 | BX041 | 1:1000 | 0.2 | | SFTPC* | Invitrogen | PA5-71842 | BX020 | 1:500 | 0.4 | | SCGB1A1* | R&D System | MAB4218 | BX043 | 1:400 | 0.5 | | β-III-Tubulin* | R&D Systems | MAB1195 | BX055 | 1:400 | 0.5 | | SCEL* | Abcepta | Abcepta | BX052 | 1:100 | 2 | | RAGE* | Abcam | ab228861 | BX028 | 1:100 | 2 | | LYVE1* | R&D Systems | AF2089 | BX025 | 1:100 | 2 | | COL1A1* | Abcam | ab88147 | BX054 | 1:100 | 2 | | CD298* | Abcam | ab167390 | BX005 | 1:100 | 2 | | CD1c* | Novus | ab156708 | BX016 | 1:50 | 4 | | SCGB3A2* | Abcam | ab240255 | BX002 | 1:400 | 0.5 | | TP63* | Abcam | ab214790 | BX006 | 1:100 | 2 | | MUC5AC* | Abcam | ab212636 | BX040 | 1:100 | 2 | | PROX1* | R&D Systems | AF2727 | BX050 | 1:200 | 1 | | CXCL4* | Peprotech | 500-P05 | BX004 | 1:200 | 1 | | | | | | | 41.1 µlAb/200µl blocking buffer / 1 slide | | | | | | | |
Note: Whenever possible barcodes and reporters are assigned to specific antibodies based on predicted antigen abundance and relative channel sensitivity in accordance with the PhenoCycler-Fusion User Guide (Akoya Biosciences). *Denotes custom-conjugated antibody. Refer to custom-conjugation section at the end of the protocol.
Incubate with Ab + Blocking Buffer Cocktail Solution
Wash slides in Coplin Jars containing dd H2O
- dd H20 #1 - 3 dips
- dd H20 #2 - immersion
0h 2m 0s
Immerse Slides in sequential Coplin jars containing the following buffers from Akoya Biosciences:
- Hydration buffer #1
0h 2m 0s - Hydration buffer #2
0h 2m 0s - Staining buffer
0h 20m 0s-0h 30m 0smax .
Carefully dry slide around tissue with a Kimwipe™.
Then pipette 190 μL Ab cocktail solution onto slide to cover tissue section.
Avoiding pipetting directly onto tissue.
Incubate slides, covered in a lightly humidified chamber, for 3h 0m 0s Room temperature .
Post Stain Wash and Multiple Fixations
Tissue slides are briefly washed in staining buffer followed by sequential fixation with paraformaldehyde, ice-cold methanol, and final fixation solution.
Incubate in sequential Coplin Jars containing:
- Staining Buffer #1 .
0h 2m 0s - Staining Buffer #2 .
0h 2m 0s - 1.6% paraformaldehyde (Diluted in Storage buffer, Akoya Biosciences from 20% stock).
0h 10m 0s - Rinse slides sequentially in 3 Coplin Jars containing 1 x PBS (3 dips each)
- Pre-chilled,
-20°C, methanol on ice0h 5m 0s - Rinse slides sequentially in 3 Coplin Jars containing 1 x PBS (3 dips each)
Prepare Final Fix 2% dilution
20 μl aliquot of Final Fix (Akoya Biosciences) diluted in 1 ml PBS
Carefully dry slide around tissue with a Kimwipe™
Then pipette 190 μL of 2% Final Fix solution onto slide to cover tissue section while avoiding pipeting directly onto tissue.
Incubate 0h 20m 0s .
Rinse slides sequentially in 3 Coplin Jars (3 dips each) PBS
Photobleaching and Storage
Slides may be stored for up to 5 days in a Coplin Jar containing Storage buffer 4°C.
In order to reduce autofluorescence from the tissue, on the day prior to imaging, lay the slide to be imaged flat in a 100 cm2 dish containing Storage Buffer (Akoya Biosciences).
Photobleach by illumination with a 200 mA, 15 watts, 1600 lumens bulb 4°C .
Reporter Plate and Experiment Design
Reporter plate design and Phenocycler-Fusion run protocols are developed using the PhenoCycler Experiment Designer Software (Akoya Biosciences).
Prepare 1x Phenocycler Buffer and Reporter Stock Solutions
Reporter Stock Solution is prepared according to guidelines in the attached Phenocycler-Fusion User Guide (Akoya Biosciences®)
Prepare 1 L of 1 x Phenocycler Buffer with Buffer additive, sufficient for 10 imaging runs. This is used in the side-car Phenocycler Buffer bottle, in the reporter stock solution and in the mounting of the flow cell. Do not filter. Store at room temperature for up to 2 weeks. Combine in clean glass container, mix well, avoid bubbles:
800mLddH2O100mL10x Buffer for Phenocycler (Akoya Biosciences)100mLBuffer Additive for Phenocycler Fusion (Akoya Biosciences)
Prepare Reporter Stock Solution
Combine, mix gently by inversion:
- 1 x Phenocycler Buffer with Buffer additive
270µL - Assay Reagent
25µL - Nuclear Stain
5µL
Total volume should equal 300 μl x N = number of cycles plus two blank cycles (# reporter plate wells) to be done in the imaging run.
Prepare Reporter Solutions for each cycle in an opaque or foil wrapped tube.
Keep Reporters on ice.
Centrifuge Reporter stock tubes briefly prior to removing aliquot.
Protect the fluorescent dye-containing Reporters from photobleaching.
Avoid introducing bubbles into the solutions.
Important to keep the reporter plate dust-free.
An imaging cycle = 4 channel detection including nuclear stain and 3 reporters, depending on filter set chosen, for example one each of ATTO550, Cy5/AF647, AF750. A multi-cycle experiment composes an imaging run.
Label an opaque microtainer for each well, each well = one imaging cycle of the imaging run. To each tube add 5 μl of each predetermined reporters, complementing the antibody barcodes to be detected in that cycle, and add the Reporter Stock Solution to make total volume 250 μl per microtainer.
Pipette 245 μl of each reporter mix into a well of a light-opaque 96 well microtiter plate (Akoya Biosciences) according to the Experimental Design developed in the experimental design app.
Also, pipette 245 μl reporter stock solution (blanks) into two wells of row H to create 2 blank wells. Seal the solution-containing wells with Akoya foil seals. The plate may be stored @ 40C for up to 14 days in accordance with the attached PhenoCycler-Fusion User Guide (Akoya Biosciences).
Multiplexed Imaging and Analysis
Image Acquistion via Phenocycler-Fusion ® (i.e., CODEX V2)
Use the Experiment Designer app, located on the PhenoCycler-Fusion Acquisition PC desktop,
to define the design of the Reporter plate and imaging parameters to be used in the experiment. Follow details provided in the attached PhenoCycler-Fusion User Guide.
Checking the Sample mask, BF overview, and FL overview, by dragging and dropping files into ImageJ after the first cycle is recommended. If major issues are observed, the run may be aborted to preserve reporters.
Do not attempt to open raw or intermediate cycle.qptiff during the imaging run.
Rapid review of the resultant image.qptiffs is performed utilizing PhenoChart 1.2.0 software. If necessary, exposure time (ms) is adjusted in the Phenocycler Experiment Designer to obtain readily detectable, specific marker signals that are below saturation. Note that after viewing qptiffs in Phenochart an_annotations.xml.lock file will appear in the image folder.
After the run return the tissue slide with attached flow cell to storage buffer 4°C. If necessary, slides can be reimaged with a new set of reporters up to 5 days post staining without loss of signal.
If necessary, warm reporter plate to Room temperature
After photobleaching in storage buffer, wash slides in PBS (250 ml; Coplin Jar)Room temperature 0h 10m 0s
After the wash, dry the bottom of the slide and around the edges of the tissue with a kimwipe.
Attach a flow cell using the flow cell assembly device (Akoya Biosciences). Center and press the flow cell onto the slide for 0h 0m 30s
Cure the flow cell adhesive by incubating the slide in 1X phenocycler buffer (Akoya biosciences)0h 10m 0s Room temperature
Fill respective Reagent reservoirs on the Phenocycler side car with DMSO, 1X phenocycler buffer, and ddH20, and place a blank flow cell in the flow cell carrier attached to the fluidics tubing.
Start an imaging run by turning on the Phenocycler fluidics system and the Phenoimager, followed by launching the Fusion software. Select Start experiment and follow the prompts.
Images were acquired utilizing the 20X (0.5 µM/pixel) objective and Fusion 1.0.8 software.
Image processing is automated via the Fusion 1.0.8 software and completed at the end of the experiment run.
Record Imaging Run MetaData
Record Slide ID, staining and imaging date in the Metadata Excel file located in the Fusion folder on the Phenocycler PC.
Likewise record the dates for biweekly Brightfield reference calibration and maintenance washes as well as monthly fluorescent and phenocycler reference calibration.
The "source storage time" = length of time (days) between slide staining date and imaging date.
The image acquisition time = length of time between time stamp on the MarkerList or CombineInputs files ( START ) and the time stamp on the Akoya whole slide scan file .qptiff ( FINISH) .
The time_since_acquisition_instrument_calibration_value is the time interval (days) between the phenocycler reference calibration date and the imaging run date.
Data backup and storage
Upload files from Phenocycler-Fusion runs to the URMC local Bluehive-Archive/archive/lungmap_lab codex folder.
Maintain directory structure for each run by creating a new folder labeled with the slide ID containing a scan# subfolder, and a temp subfolder within the scan folder.
Include the following files in the upload:
Slide folder:
Scan Folder:
A. (.xpd) file (Phenocycler Experiment designer)
B. Akoya whole slide scan .qptiff
C. The ome.tiff generated via bfconvert (See step 17)
Temp folder:
9. CombineInputs
ii. Coverslip Mask
iii. qptiff raw files for each cycle
iv. qptiff.intermediate files for each cycle
v. FocusMap
vi. FocusTable
vii. Label
viii. MarkerList
ix. Overview BF
x. Overview FL
xi. SampleMask
Image Analysis and Segmentation
Analysis of processed image.qptiff files is performed utilizing QuPath (See Reference 3).
Cell segmentation based on (DAPI) stained nuclei is performed utilizing the respective StarDist extension (See Reference 4) in QuPath.
Cell detection measurements (e.g. X,Y coordinates, fluorescent intensity data for all channels) are saved as a text file, opened in Excel as a tab delimited file, and resaved as a CSV (comma delimited) file.
Cell segmentation masks are exported from QuPath running the following ExpCell script within Script Editor:
import qupath.lib.images.servers.LabeledImageServer
import qupath.lib.roi.RectangleROI
import qupath.lib.scripting.QP
def imageData = getCurrentImageData()
def project = getProject()
def server = imageData.getServer()
// Define output path (relative to project)
def name = GeneralTools.getNameWithoutExtension(imageData.getServer().getMetadata().getName())
def pathOutput = buildFilePath(PROJECT_BASE_DIR, 'export')
mkdirs(pathOutput)
// Define output resolution
double requestedPixelSize = server.getPixelCalibration().getAveragedPixelSize()
// Convert to downsample
double downsample = requestedPixelSize / imageData.getServer().getPixelCalibration().getAveragedPixelSize()
println('Downsample factor : '+downsample)
// Create an ImageServer where the pixels are derived from annotations
def labelServer = new LabeledImageServer.Builder(imageData)
.backgroundLabel(0, ColorTools.BLACK) // Specify background label (usually 0 or 255)
.downsample(downsample) // Choose server resolution; this should match the resolution at which tiles are exported
.useCells()
.useInstanceLabels() //Export as unique label
.multichannelOutput(false) // If true, each label refers to the channel of a multichannel binary image (required for multiclass probability)
.build()
annotation = getSelectedObject()
//Export the mask
def region = RegionRequest.createInstance(labelServer.getPath(), downsample, annotation.getROI())
def outputPath = buildFilePath(pathOutput, 'labels_mask.ome.tiff')
writeImageRegion(labelServer, region, outputPath)
resetSelection()
Vascular smooth muscle and airway epithelial features are annotated and classified by thresholding the ACTA2 and PanCK signals utilizing the Pixel classifier in QuPath.
Annotation masks are exported from QuPath running the following ExpCell script within Script Editor:
def imageData = getCurrentImageData()
// Define output path (relative to project)
def outputDir = buildFilePath(PROJECT_BASE_DIR, 'export')
mkdirs(outputDir)
def name = GeneralTools.getNameWithoutExtension(imageData.getServer().getMetadata().getName())
def path = buildFilePath(outputDir, name + "-labels.png")
// Define how much to downsample during export (may be required for large images)
double downsample = 8
// Create an ImageServer where the pixels are derived from annotations
def labelServer = new LabeledImageServer.Builder(imageData)
.backgroundLabel(0, ColorTools.WHITE) // Specify background label (usually 0 or 255)
.downsample(downsample) // Choose server resolution; this should match the resolution at which tiles are exported
// Choose output labels (the order matters!)
.addLabel('Airway', 1)
.addLabel('Vasculature', 2)
.addLabel('Other', 3)
.multichannelOutput(false) // If true, each label refers to the channel of a multichannel binary image (required for multiclass probability)
.build()
// Write the image
writeImage(labelServer, path)
Object Data is exported as a GeoJSON file.
Conversion of .qptiff files to ome.tiff for upload to Omero utilizing bfconvert
Download Command Line Tools from https://www.openmicroscopy.org/bio-formats/downloads/ https://www.openmicroscopy.org/bio-formats/downloads/
Execute the following in command prompt:
C:>Set BF_MAX_MEM=10240M
C:>bfconvert -no-sas -series 0 -compression LZW -pyramid-resolutions 5 -pyramid-scale 2 INPUT.qptiff OUTPUT_ome.tiff
Upload the resulting ome.tiff file to the CODEX folder in Omero and the respective tissue donor subfolder (e.g. D265) utilizing the Omero importer.
Custom antibody conjugation
Custom Antibody Conjugation is performed as described dx.doi.org/10.17504/protocols.io.3fugjnw. dx.doi.org/10.17504/protocols.io.3fugjnw.
For antibodies containing sodium azide (0.05-0.1%) or trehelose (5%), buffer exchange was performed utilizing Zeba Spin Desalting columns 7K MWCO (89890, 2ml, Thermoscience) equilibrated in PBS in accordance with the manufacturer's recommendations.
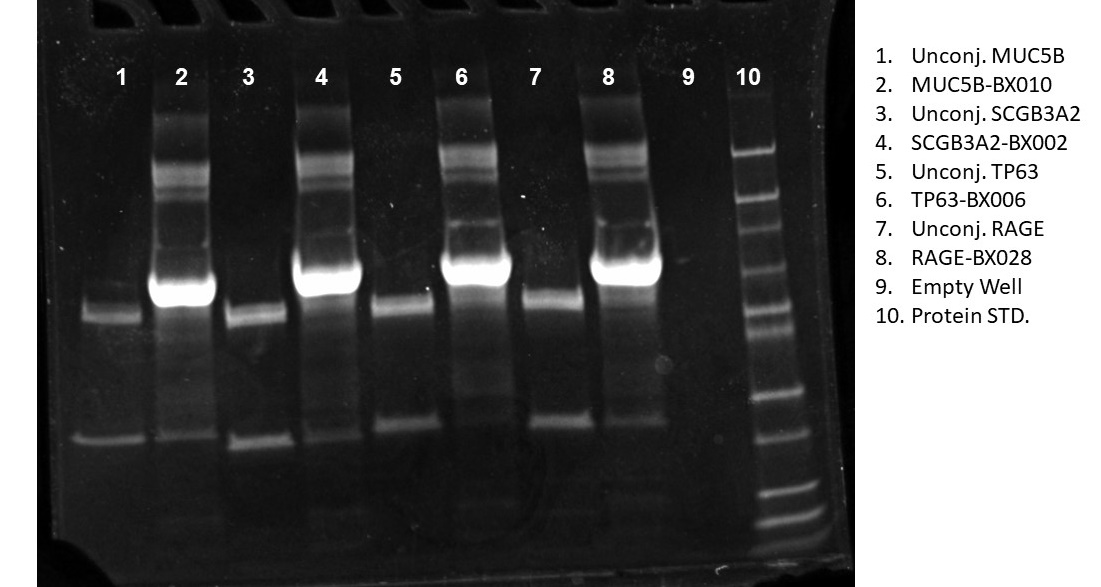
Success of Antibody-Barcode chemical conjugation is determined by resolving unconjugated and conjugated Ab's on BioRADTMs MiniProtean TGX Gel 4-15% Bis-Tris Protein Gels in accordance with Guidelines in the Phenocycler-Fusion User Guide (Akoya Biosciences®, Malborough, MA).
H&E staining Post Phenocycler-Fusion
It is useful to stain the tissue sections with hematoxylin & eosin following the multiplexed fluorescence imaging in order to match histology with antibody staining. We have stained lung sections with an aqueous H&E stain as described dx.doi.org/10.17504/protocols.io.kqdg397yeg25/v1 but this resulted in an overall rather grey stain and so we have opted instead to remove the flow cell by soaking in xylene prior to H&E staining of lung sections as described in Step 19.1.
After all MxF imaging is completed for the slides, remove the flow cell covered slides from storage buffer (Akoya Biosciences), wipe dry with Kimwipe and, utilizing a vacuum-suction apparatus similar to what is described in dx.doi.org/10.17504/protocols.io.kqdg397yeg25/v1, remove buffer from the flow cell.
- Incubate slides in a Coplin Jar containing Xylene for up to
4h 0m 0sin order to weaken or dissolve the flow cell adhesive. - Carefully pry flow cell off the slide with razor blade or preferably a thin spatula.
- Proceed with H&E staining according to the standard protocol described here dx.doi.org/10.17504/protocols.io.36wgqjnq3vk5/v4 beginning at Step 5.