3D-correlative FIB-milling and Cryo-ET of Autophagic structures in Yeast Cells
Cristina Capitanio, Anna Bieber, Philipp S Erdmann, Brenda A Schulman, Florian Wilfling, Wolfgang Baumeister
Abstract
This protocol describes how to plunge-freeze yeast on EM grids and how to target autophagic structures by combining cryo confocal fluorescence data to FIB-milling and tomogram acquisition
Steps
YEAST CELL CULTURE
Prepare and starve yeast cells before plunging:
Inoculate Yeast from overnight cultures in YPD medium to an OD600 of 0.15.
Let cultures grow at 30°C, 200 rpm, till OD600 is 0.8. At this point, switch to starvation medium SD-N.
Incubate cells in starvation medium at 30°C for a time span of 0.5 - 3 hours.
Before plunging, add 1 μm beads to the cell solution, at a dilution of 1:20.
The beads are necessary in the next steps to perform 3D-correlation on the grid.
PLUNGE FREEZING
Prepare Vitrobot and grids for plunging:
-
Set up Vitrobot as follows: Blotforce = 8, Blottime = 10s, room temperature.
-
Plasma clean grids (200 Mesh Cu SiO2 R1/4, Quantifoil) for 30 s, just before plunging.
Apply 4 µl of starved cell solution with beads on the grid, blotted and plunged in ethane-propane with a Vitrobot Mark IV and plunge.
Store the grids in a grid box in liquid nitrogen.
CRYO-CONFOCAL FLUORESCENCE MICROSCOPY
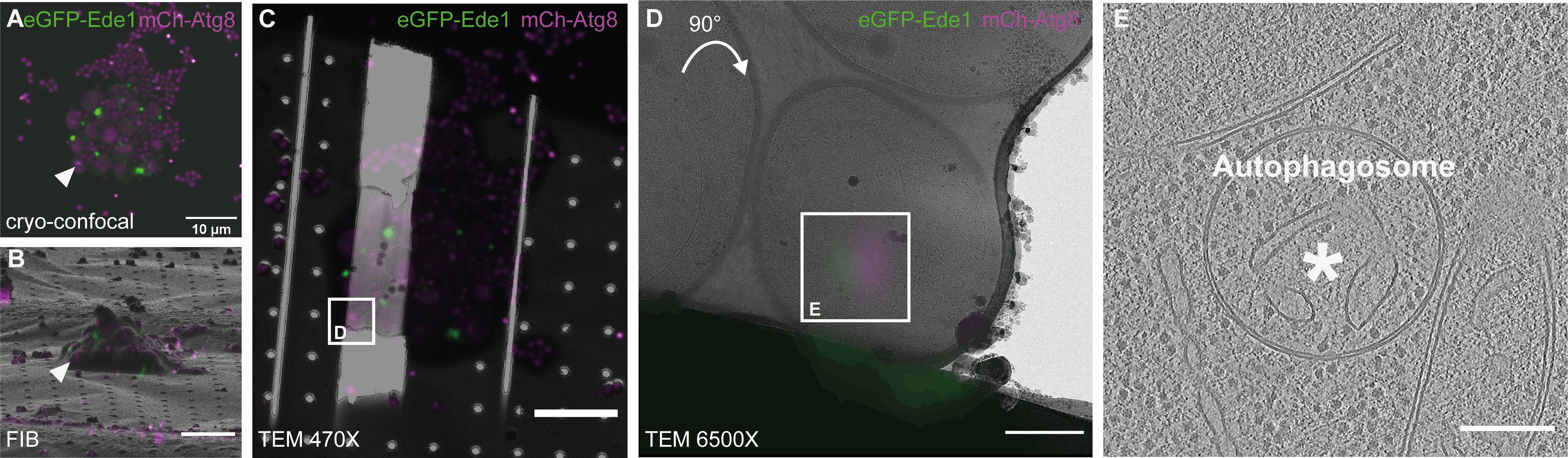
Take fluorescence stacks of target cells on a cryo-confocal fluorescence microscope for correlative focused ion beam milling and cryo-ET:
Clip/mount grids on modified autogrids with cut-out for FIB-milling and load the grid into the cryo-fluorescence microscope.
Acquire a fluorescence and transmitted light grid overview (e.g. using a spiral scan). If parts of the grid are out of focus, acquire a focus map by autofocusing in ~15 points spread over the grid and take acquire a second overview using the focus map.
Find grid squares suitable for FIB milling, which fulfill the following criteria:
- The square contains 1-2 clumps of 4-8 yeast cells close to the center of the grid square
- The cells show at least one fluorescence punctum indicating the presence of an autophagic structure.
- The square has at least 8 separate fiducial beads spread across the grid square
- The square is located in a position accessible in the FIB/SEM instrument, i.e., no closer than 4 grid squares to the edge of a 200 mesh grid. Mark the identified grid squares on the overview to allow later identification in the FIB/SEM instrument.
For each grid square, acquire a stack in the appropriate fluorescence channels, with pixel size and focus step appropriate for later deconvolution. On the Leica SP8 system with an NA 0.9 objective, we usually use a xy pixel size of 85 nm and a focus step of 300 nm.
Deconvolve the fluorescence stacks with appropriate software, e.g. using Huygens deconvolution software, and reslice them to isotropic pixel size e.g. with the 3DCT for correlation.
CORRELATIVE FOCUSED-ION BEAM MILLING
Mill lamellas containing the fluroescently labelled target structures for tomogram acquisition:
Load grid into the FIB/SEM shuttle with the cutout facing upwards. Apply a protective organometallic layer (CpMePtMe3) on the grid using the Gas Injection System (GIS).
Acquire an SEM overview image of the whole grid. Using holes or grid bar junctions as landmarks, perform a 2D correlation with the grid overview acquired in the lfluorescence microscope to find all target squares.
Visit each target grid square and save its position at eucentric height. Take high-resolution SEM and ion beam (IB) images at the milling angle for correlation. On a Scios/Aquilos FIB/SEM instrument, we usually use a milling angle of 18°, corresponding to a lamella pre-tilt of 11° relative to the grid. Taking the images of all the squares first allows for an easier parallelization of image registration and FIB milling and thus a more efficient use of instrument time.
For each square, perform 3D correlation with the 3D correlation toolbox (3DCT) as follows:
- Load the resliced deconvolved fluorescence stack as first, and the ion beam image as second image. For colocalization experiments, load additional fluorescence stacks as extra channels. Display fluorescence data as maximum intensity projections.
- Identify and mark four corresponding beads in the fluorescence and ion beam image. Refine the bead positions in the fluorescence data by performing an x,y,z gaussian fit, then click "Correlate" to calculate the initial registration.
- Click further beads in the fluorescence image, refine their 3D positions and predict their positions in the IB image by clicking "Correlate". Click the correct bead positions in the IB image and repeat.
- Iterate step 3.4.3 until reaching a consistent registration with 8-14 beads.
- Select the target fluorescence punctum, refine its 3D position with a Gaussian fit and predict its position in the ion beam. The red marker displayed on the ion beam image indicates the target position for the lamella.
Mill the lamella at the target position, milling simultaneously from above and below. The ion beam currents and target thicknesses of our usual milling steps (2xrough and 2xfine) are:
- 0.3 nA - 1.2 µm
- 0.1 nA - 800 nm
- Optional: Add stress release cuts in the grid square on both sides of the lamella.
- 50 pA - 250 nm
- 30 pA - 100-150 nm, or until transparency at SEM voltage of 3 kV.
After milling, acquire an SEM and IB image of the grid square with the final lamella and perform 3D correlation with the pre-milling fluorescence data as described in step 3.4 to verify that the lamella is at the correct position, and to identify the target position for tomogram acquisition.
After milling all lamellas, take an SEM overview of the whole grid to allow easy recognition of all lamellas in the TEM.
CORRELATIVE CRYO-ET TOMOGRAM ACQUISITION
Acquire tomograms of target structures in correlatively milled lamellas:
Load grids into the TEM with the previous milling direction perpendicular to the tilt axis.
Acquire an overview image of each grid square containing a lamella, and a montage of each lamella.
To determine positions for tomogram acquisition, register the pre-milling fluorescence data with the TEM grid square overview using 3DCT as described in step 3.4. Check the identified positions in the lamella overview to center the target structure during tilt series acquisition.
Run tilt-series at the correlated positions, using e.g. batch acquisition in SerialEM Navigator. We usually use a dose-symmetric tilt scheme with 2° steps starting from a lamella pretilt of 10° to +70° and -50°. Images are acquired in dose-fractionation mode and a final target dose of 120 e-/Å2.