isolation and extraction of plant nuclei in plug
Benoît Vacherie, Karine Labadie
Abstract
Method for isolation and extraction of plant cell nuclei.
Protocol for obtaining UHMW DNA (> 150kb) allowing the production of optical cards with Bionano technology.
Steps
preparation of reagents
NIB Buffer : 200 ml : freshly prepared
| A | B |
|---|---|
| Tris ph8 | 10 mM |
| EDTA | 10 mM |
| KCl | 80 mM |
| Sucrose | 0.5 M |
| PVP 40 | 2 % |
| Spermine | 1 mM |
| Spermidine | 1 mM |
| H2O | qsp 200 ml |
Adjust the Ph to 9.4 then filter at 0.22 µm
NIBT Buffer : 160 ml
| A | B |
|---|---|
| NIB Buffer | 160 ml |
| Triton X100 | 0.5% |
NIBTM Buffer : 40 ml
| A | B |
|---|---|
| NIBT Buffer | 40 ml |
| 2-Mercaptoethanol | 0.75 % |
Cell suspension Buffer : Can be stored for 1 year at 4°C.
| A | B |
|---|---|
| Tris ph8 | 10 mM |
| EDTA | 50 mM |
| NaCl | 2 mM |
| H2O | Qsp 100ml |
Lysis Buffer : Can be stored for 1 year at RT.
| A | B |
|---|---|
| EDTA 0.5M | 100 ml |
| N-Lauroylsarcosine | 1 % |
Nuclei isolation
Putting a mortar in ice
Cool a mortar/pestle with liquid nitrogen until the bubbling stops.
Place a beaker in ice and add a magnetic stirrer.
Add 20 ml of NIBTM (10 ml/g of leaves) and stir gently
20mL
Filter the mixture into a 50ml tube through autoclaved filters (2 cheese cloth + 2 Mira cloth) on a funnel (squeeze the filters at the end of filtration to recover more of the solution containing the nuclei).
Filter through a 40µm cell stariner into a new 50ml tube.
Remove the supernatant and gently resuspend the pellet in ice (use a brush if the pellet does not recover).
Add 20ml of cold NIBTM
20mL
Filter the supernatant through a 40 µm cell sieve into a new 50 ml tube.
Remove the supernatant with a pipette and resuspend the pellet in an appropriate volume of cell suspension buffer :
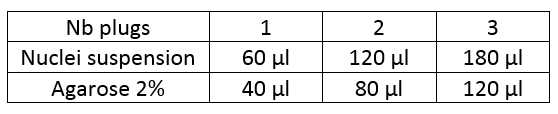
60µl / plug
Adjust the number of plugs to be made according to the size of the pellet.
Embedding in agarose
Put a CHEF Disposable Plug Molds on ice
Melt agarose at 70°C for 5 min then equilibrate at 43°C for 5 min
0h 5m 0s 70°C
0h 5m 0s 43°C
Preheat the nuclei suspension to 43°C for 3min and then add the appropriate amount of 2% agarose (see table).
Mix gently with a wide-bore tip, avoiding bubbles.
0h 3m 0s 43°C
Proteinase K digestion
Prepare a fresh proteinase K digestion solution by mixing 200 μl of proteinase K enzyme (20mg/ml) with 2.5 ml of lysis buffer in a 50 ml tube.
200µL 2.5mL
Transfer plugs to the 50ml tube containing Proteinase K digestion solution.
Incubate in thermomixer for 2 hours at 50 °C with intermittent mixing
Mixing cycle : 10 seconds at 450 rpm followed by 10 minutes at 0 rpm
2h 0m 0s 50°C
Screw a sieve caps onto the tube and empty the solution. Change the proteinase K Solution bath as before.
Incubate in thermomixer overnight at 50 °C with intermittent mixing
Mixing cycle : 10 seconds at 450 rpm followed by 10 minutes at 0 rpm
50°C
RNase Digestion
Empty the tube using a vent cap.
Rinse the plugs 3 times with 10ml of wash buffer.
Wash 2 times with 10ml wash buffer for 15 min at RT with gentle agitation (15 rpm) on a horizontal platform mixer.
15rpm
Rinse the plugs 3 times with 10ml of TE 10:5
Add 2.5ml of TE 10:5 and 50 µl of Rnase Solution
Incubate 1hour at 37°C with intermittent mixing
1h 0m 0s 37°C
Rinse the plugs 3 times with 10ml of Wash Buffer.
***NB : The plugs can be stored at 4°C in a wash buffer at this stage***
Agarase treatment
Wash 4 times with 10ml wash buffer for 15 min at RT with gentle agitation (15 rpm) on a horizontal platform mixer.
15rpm
Transfer the plug to a 1.5 ml tube with a sterile spatula
Melt the plug in a water bath at 70°C for 2 minutes
0h 2m 0s 70°C
Transfer the tube to a water bath at 43°C for 5 minutes
0h 5m 0s 43°C
Add 2µl of agarase and mix gently by rotating with the tip.
Incubate 45 minutes at 43 °C
0h 45m 0s 43°C
Dialysis
Place 10 ml of 1x TE Buffer in a 6 cm Petri dish.
Float a 0.1 μm dialysis membrane on the surface of the 1x TE Buffer. Place a cover on the Petri dish and let the membrane equilibrates for 15 minutes.
0h 15m 0s
Deposit the entire sample in the centre of the membrane using a wide-bore tip.
Place cover on the Petri dish and let the sample dialyze for 45 minutes at room temperature.
0h 45m 0s Room temperature
Transfer DNA to a 1.5 ml microfuge tube with a Wide Bore Tip.
Allow the DNA to resuspend overnight at RT then 2 days at 4°C before performing quality control.
Room temperature
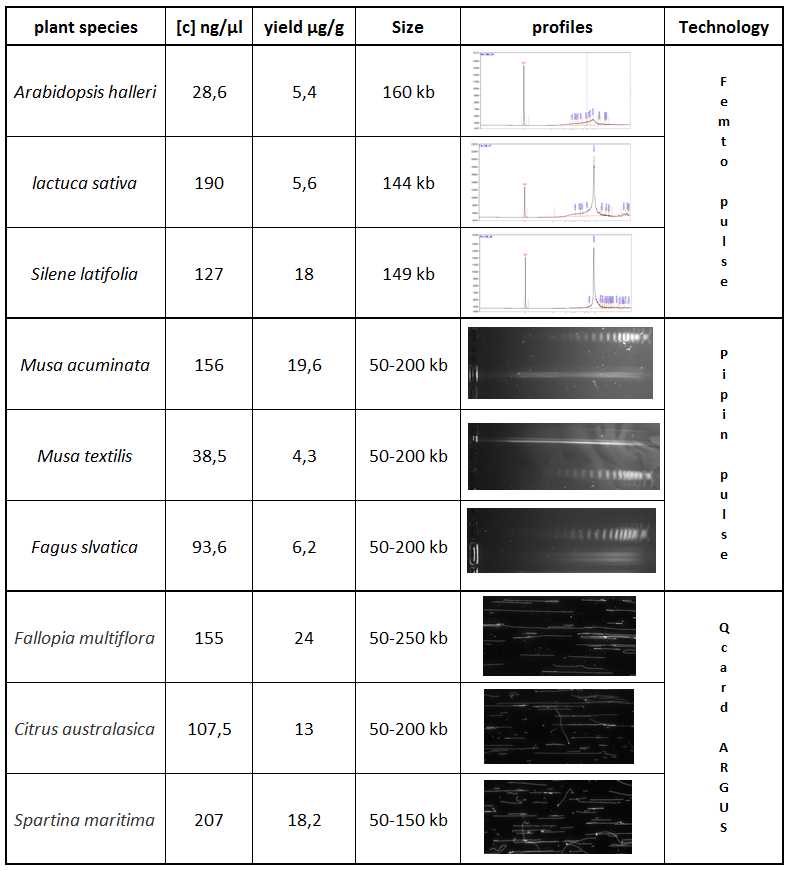
Sample QC
Quantify your sample with a Qubit HS .
NB : Before quantification, sonicate the DNA aliquot for 10 min to obtain a more reliable result
Visualise 1 µL of sample to estimate the molecular weight. ( Tapestation or/and pipin pulse or/and Femto pulse )