cDNA library preparation using ARTIC v3 primers and NEB UDI UMI adaptors for NGS
Livia C T Scorza, Trevor Ho, Nahuel Manzanaro Moreno, Marion Walker, Nick Gilbert
Abstract
This is a cDNA library preparation protocol with the aim to quantify SARS-CoV-2 in wastewater and potentially detect its different variants using Next Generation Sequencing (Illumina).
In this protocol, we amplified cDNA from viral RNA extracted from wastewater.
The main steps involve:
- Amplification of cDNA with ARTIC v3 primers
- Ligation of amplified cDNA to NEB UDI UMI adaptors
- PCR enrichment of the libraries
- Pooling libraries together for sequencing
The further analysis pipeline used to perform assembly and intra-host/low-frequency variant calling is the nf-core/viralrecon: https://nf-co.re/viralrecon
This protocol is still under revision
Steps
cDNA synthesis
| A | B |
|---|---|
| NTC | No-template control |
| EPC | External positive control |
| Sample | wastewater sample |
It is recommended to add as templates a positive and a negative control.
For cDNA synthesis, refer to
NEBNext® ARTIC SARS-CoV-2 Library Prep Kit (Illumina®) NEB #E7650S/L NEB ARTIC E7650.pdf
Chapter 2 Standard Protocol with cDNA Amplicon and Ligation Bead Cleanups (Three clean-up steps)
- Follow steps 2.1 to 2.3
After cDNA clean up, you can run 1 µL of the cDNA, diluted 1:10 on Agilent TapeStation with HS D1000 Tapes to look at the size distribution.
If you are confident the sizes would be correct, just use Qubit HS dsDNA protocol to quantify the input DNA amount.
Store the amplified cDNA at -20 °C.
Ligate amplified cDNA to NEB UDI UMI adaptors
For choosing adaptors, follow the following manual:
NEBNext® Multiplex Oligos for Illumina® - Unique Dual Index UMI Adaptors DNA Set 1
NEB #E7395S
"Designed for use in library prep for DNA, ChIP DNA and RNA (but not Small RNA), the NEBNext Unique Dual Index UMI (Unique Molecular Identifier) Adaptors enable high-efficiency adaptor ligation and high library yields.
These adaptors contain all necessary sequences for sequencing on the Illumina platform, and are compatible with PCR-free applications and sample pooling prior to PCR amplification".
Refer to tables 1-4 for information on index sequences
Check Index Pooling guidelines in the manual mentioned above (NEB #E7395S )
End Prep
Follow the step 2.4 of NEBnext End Prep protocol in the NEB kit for SARS-CoV-2 detection manual ( E7650)
Adaptor dilution and ligation
In this step we use parts of both protocols (E7395 and E7650)
-
Adaptors with UMI are used
-
A smaller reaction volume is used following that of the ARTIC kit
Adaptor dilution (optional depending on amount of input DNA)
In this step the cDNA quantification information is detrimental. Depending on the amount of DNA input, adaptor dilution is a necessary step.
"If DNA input is ≤ 100 ng, dilute NEBNext Adaptor"
Please refer to NEBNext® Ultra™ II DNA Library Prep Kit for Illumina®
NEB #E7645S/L, #E7103S/L NEBNext Ultra II DNA library prep for Illumina E7103-E7645.pdf
Step 2. "Adaptor Ligation - Determine whether adaptor dilution is necessary"
Adaptor ligation
Use NEBNext® Ultra™ II DNA Library Prep Kit for Illumina® NEB #E7645S/L, #E7103S/L as a guide
The E7650 protocol uses NEBNext adaptors for Illumina which are truncated and must be PCRed to add the full P5 and P7 sequences.
Those NEBNext adaptors also need the USER enzyme because the adaptors need to be cleaved prior to the PCR step.
The E7395 protocol does not require this.
Mixing together these features the ligation recipe was rewritten as follows:
| A | B | C |
|---|---|---|
| COMPONENT | VOLUME (µL) | 10.4 |
| End Prep Reaction Mixture | 30 | - |
| Diluted or undiluted NEBNext UMI Adaptors for Illumina | 1.25 | - |
| (red) NEBNext Ultra II Ligation Master Mix | 15 | 156 |
| (red) NEBNext Ligation Enhancer | 0.5 | 5.2 |
| Total Volume | 46.75 |
modified from step 2.2 of NEBNext® Ultra™ II DNA Library Prep Kit for Illumina® NEB #E7645S/L, #E7103S/L
Follow to steps 2.3 and 2.4 of the aforementioned manual (NEBNext® Ultra™ II DNA Library Prep Kit for Illumina® NEB #E7645S/L, #E7103S/L)
Samples can be stored at - 20 overnight
Cleanup of Adaptor-ligated DNA
As we know most amplicons are 400 bp in size, it is not necessary to use the size selection protocol.
Rather, we move to section 3B Cleanup of Adaptor-ligated DNA without size selection for all samples
Protocol: NEBNext® Ultra™ II DNA Library Prep Kit for Illumina® NEB #E7645S/L, #E7103S/L
PCR enrichment
Follow on to Step 4. PCR Enrichment of Adaptor-ligated DNA
Protocol: NEBNext® Ultra™ II DNA Library Prep Kit for Illumina® NEB #E7645S/L, #E7103S/L
Determine the number of cycles in your PCR reaction:
Proceed to Step 4.1. PCR Amplification
From NEBNext® Ultra™ II DNA Library Prep Kit for Illumina® NEB #E7645S/L, #E7103S/L
PCR enriched library clean up
Follow on to Step 5 of the same manual
Checking library size distribution and library quantification
After clean up, run 0.5 µL of the cDNA (diluted 1:10 4.5 μL of water) on a Agilent TapeStation with HS D1000 Tapes to look at the size distribution.
We recommend running a few randomly selected libraries to confirm size distirbution.
If this the first time doing it and we see consistent aberrant patterns showing up, then it will be necessary to run all libraries to judge how widespread the issue is.
Below is an example of expected size distribution
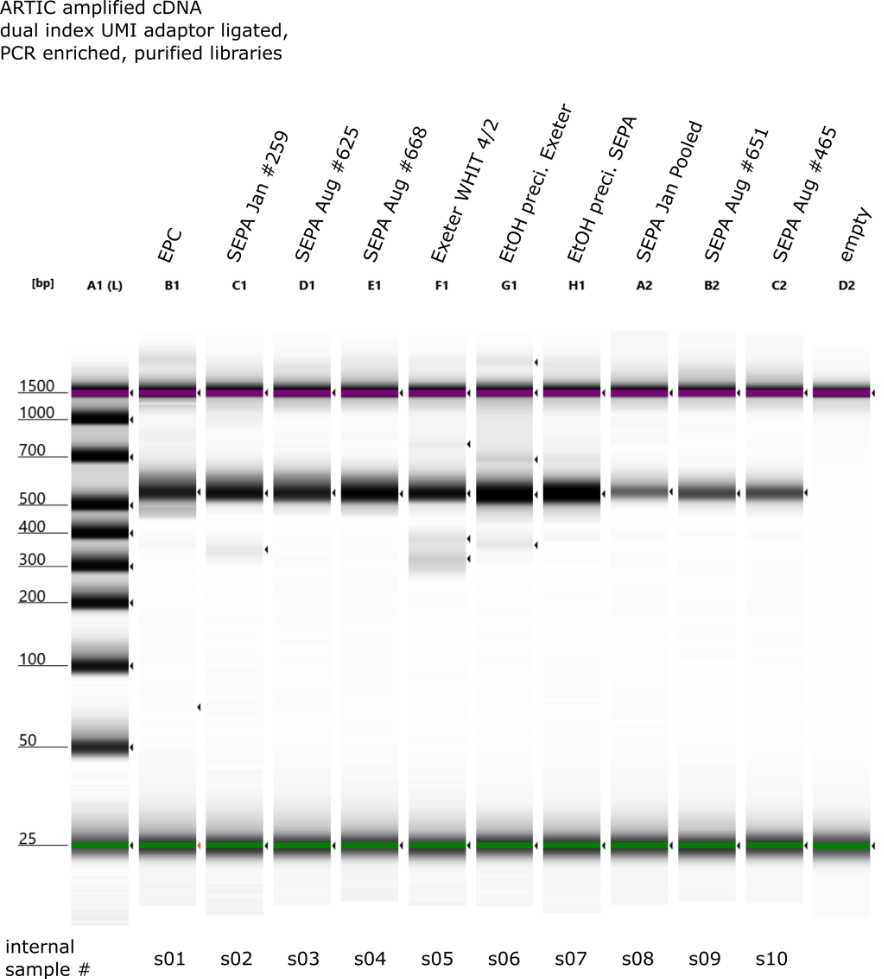
Use a quantification method such as Qubit HS dsDNA protocol to quantify the input DNA amount.
DNA quantification here is very important
Pooling the libraries for sequencing
The samples that are going to be sequenced have to be pooled together in equimolar concentrations.
Example of how samples were prepared
| A | B | C | D | E | F | G | H | I | J |
|---|---|---|---|---|---|---|---|---|---|
| Sample ID | Total library DNA amount (µg) | Remaining volume (µL) | Concentration (ng/µL) | Volume ratio to reach lowest concentration | Scaled volume to mix (µL)* | Rounded volume (µL) | DNA amount in mixed volume (ng) | Mixed DNA concentration (ng/µL) | Mixed DNA concentration (nM) |
| s001 | 1.9942 | 29.5 | 67.6 | 0.4452662722 | 6.2337278107 | 6.23 | 421.148 | ||
| s002 | 2.22725 | 29.5 | 75.5 | 0.3986754967 | 5.5814569536 | 5.58 | 421.29 | ||
| s003 | 2.03845 | 29.5 | 69.1 | 0.4356005789 | 6.0984081042 | 6.1 | 421.51 | ||
| s004 | 2.65795 | 29.5 | 90.1 | 0.3340732519 | 4.6770255272 | 4.68 | 421.668 | ||
| s005 | 2.1594 | 29.5 | 73.2 | 0.4112021858 | 5.7568306011 | 5.76 | 421.632 | ||
| s006 | 3.363 | 29.5 | 114 | 0.2640350877 | 3.6964912281 | 3.7 | 421.8 | ||
| s007 | 3.5105 | 29.5 | 119 | 0.2529411765 | 3.5411764706 | 3.54 | 421.26 | ||
| s008 | 0.88795 | 29.5 | 30.1 | 1 | 14 | 14 | 421.4 | ||
| s009 | 1.1859 | 29.5 | 40.2 | 0.7487562189 | 10.4825870647 | 10.48 | 421.296 | ||
| s010 | 1.1918 | 29.5 | 40.4 | 0.745049505 | 10.4306930693 | 10.43 | 421.372 | ||
| Total volume | 70.5 | 4214.376 | 59.7783829787 | 164.6787409882 |
Scaled volume to mix was determined by looking at the sample with lowest concentration (s008), to which we should add 14 µL first (out of 29.5 µL)

