Using Nanocompore to Identify RNA Modifications from Direct RNA Nanopore Sequencing Data
Logan Mulroney, Logan Mulroney, Ewan Birney, Ewan Birney, Tommaso Leonardi, Tommaso Leonardi, Francesco Nicassio, Francesco Nicassio
Abstract
RNA modifications can alter the behavior of RNA molecules depending on where they are located on the strands. Traditionally, RNA modifications have been detected and characterized by biophysical assays, mass spectrometry, or specific next-generation sequencing techniques, but are limited to specific modifications or are low throughput. Nanopore is a platform capable of sequencing RNA strands directly, which permits transcriptome-wide detection of RNA modifications. RNA modifications alter the nanopore raw signal relative to the canonical form of the nucleotide, and several software tools detect these signal alterations. One such tool is Nanocompore, which compares the ionic current features between two different experimental conditions (i.e., with and without RNA modifications) to detect RNA modifications. Nanocompore is not limited to a single type of RNA modification, has a high specificity for detecting RNA modifications, and does not require model training. To use Nanocompore, the following steps are needed: (i) the data must be basecalled and aligned to the reference transcriptome, then the raw ionic current signals are aligned to the sequences and transformed into a Nanocompore-compatible format; (ii) finally, the statistical testing is conducted on the transformed data and produces a table of p -value predictions for the positions of the RNA modifications. These steps can be executed with several different methods, and thus we have also included two alternative protocols for running Nanocompore. Once the positions of RNA modifications are determined by Nanocompore, users can investigate their function in various metabolic pathways. © 2023 The Authors. Current Protocols published by Wiley Periodicals LLC.
Basic Protocol : RNA modification detection by Nanocompore
Alternate Protocol 1 : RNA modification detection by Nanocompore with f5c
Alternate Protocol 2 : RNA modification detection by Nanocompore using Nextflow
INTRODUCTION
RNA modifications have been demonstrated to be important for many cellular processes, such as splicing, translation, and RNA stability (Anreiter, Mir, Simpson, Janga, & Soller, 2021). To date, there are ∼165 known naturally occurring RNA modifications (Boccaletto et al., 2018), and disruptions to physiological regulation of modification pathways are linked to diseases such as cancer (Anreiter et al., 2021; Barbieri & Kouzarides, 2020). Traditionally, RNA modifications have been detected and identified using biophysical assays, mass spectrometry, and next-generation sequencing (NGS; Helm & Motorin, 2017). However, the NGS methods are limited to a small set of abundant modifications, mass spectrometry provides limited site-specific information, and biophysical assays have both of these limitations (Helm & Motorin, 2017). Oxford Nanopore Technologies (ONT) has developed a technique to directly sequence RNA molecules without the need for a cDNA intermediate. This technique, direct RNA nanopore sequencing (DRS), has the potential to detect all possible RNA modifications with near–single nucleotide precision because they are not erased during cDNA synthesis (Garalde et al., 2018; Workman et al., 2019). Modified nucleotides systematically alter the ionic current relative to the unmodified form of the nucleotide (Smith, Jain, Mulroney, Garalde, & Akeson, 2019), and these ionic current alterations can be used to determine the presence and position of modifications in RNA strands (Begik et al., 2021; Leger et al., 2021; Pratanwanich et al., 2021; Stoiber et al., 2017).
At the time of this writing, there are at least fifteen software tools that aim to identify RNA modifications from DRS data (Furlan et al., 2021). These tools fall into two basic categories: tools that detect RNA modifications from systematic nucleotide miscalls introduced by the Oxford Nanopore Technologies (ONT) basecalling algorithm (Abebe et al., 2022; Jenjaroenpun et al., 2021; Liu, Begik, & Novoa, 2021; Parker et al., 2020) and tools that detect RNA modifications from the raw ionic current data (Begik et al., 2021; Gao et al., 2021; Leger et al., 2021; Lorenz, Sathe, Einstein, & Yeo, 2020; Maier, Gressel, Cramer, & Schwalb, 2020; Pratanwanich et al., 2021). Additionally, some tools detect modifications by comparing the data from two different conditions (Abebe et al., 2022; Begik et al., 2021; Leger et al., 2021; Liu et al., 2021; Parker et al., 2020), while others detect RNA modifications directly from the samples of interest (Gao et al., 2021; Lorenz et al., 2020; Maier et al., 2020). Furthermore, some tools increase their specificity and sensitivity by limiting the scope of detection to a subset or a single type of RNA modification (Gao et al., 2021; Liu et al., 2019; Lorenz et al., 2020). Nanocompore is a software tool that detects RNA modifications by comparing the raw ionic current data from two different sample conditions (Leger et al., 2021). Comparing the ionic current features from two samples controls for any changes in the signal that are not due to the experimental conditions under investigation and may lead to misinterpretation, such as alterations to the ONT sequencing platform. Nanocompore has also been demonstrated to be among the tools with the highest specificity for detecting RNA modifications (Acera Mateos et al., 2022; Leger et al., 2021). This high specificity, combined with a flexible experimental design, makes Nanocompore an ideal tool for detecting RNA modifications from DRS data.
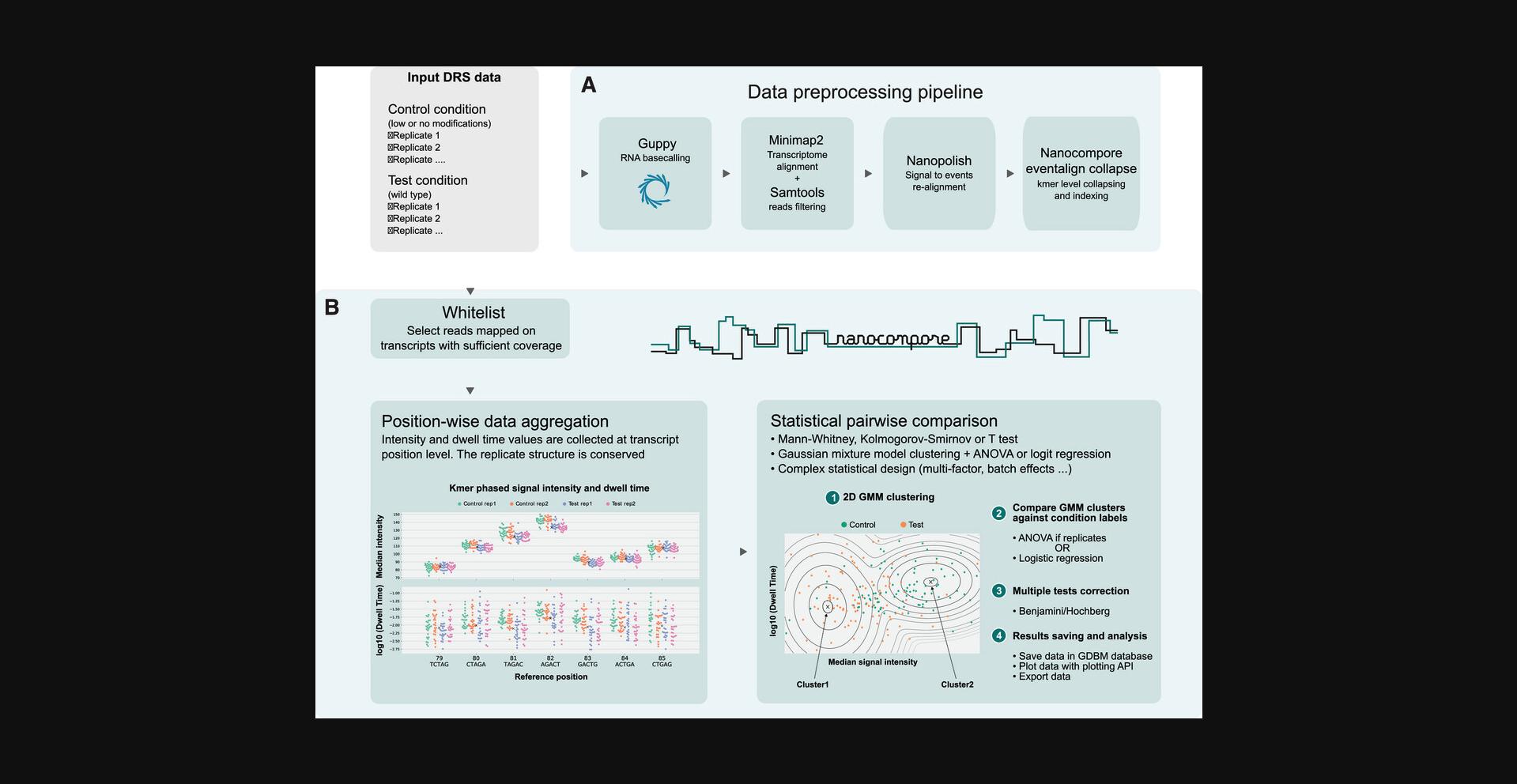
The computational analysis protocol described herein (see Basic Protocol) shows how to extract features of the raw ionic current signal from DRS to detect RNA modifications on RNA strands using Nanocompore (Leger et al., 2021; Fig. 1). The first step of the protocol is to pre-process the data into a form where the pairwise statistical tests can be performed. This involves basecalling the data using the ONT basecaller, aligning the reads to the reference transcriptome with minimap2, aligning the ionic current data to the sequence (also known as resquiggling) using nanopolish, and transforming the resquiggled data output with Nanocompore. The second step of the protocol performs the position-by-position pairwise comparison between two sample conditions to detect RNA modifications using Nanocompore. The output of Nanocompore is a tab-separated file (.tsv) that contains summary information and the results for each statistical test for each position in the reference transcriptome file. This .tsv file can be filtered on the resulting p -values to determine positions that are significantly different between the two conditions. Additionally, we introduce two Alternate Protocols that can be followed to achieve the same results, but with different time and installation considerations.
The first step to understanding is observation, and Nanocompore provides a technique to observe RNA modifications in a way that was previously challenging. The results from this protocol can be used to determine the positions of one or more types of RNA modifications in a transcriptome of interest. These observations can be used to further understand the functions of many different RNA modifications, such as in RNA stability (Boo & Kim, 2020), RNA splicing (Martinez & Gilbert, 2018), immunity (Cui et al., 2022), and many other cellular pathways.
STRATEGIC PLANNING
The comparative approach of Nanocompore permits experimental design flexibility, but the reference sample should be carefully considered. The key component for successful RNA modification detection with Nanocompore is the reference sample. The reference sample is derived from an experimental condition with a reduced abundance of the target modifications compared with the test sample. The most common example is comparing DRS data from a sample of interest where an enzyme that catalyzes a specific RNA modification (writer) is down-modulated by knock-down or genetically removed (knock-out) (Leger et al., 2021; Price et al., 2020). This experimental design detects the targeted enzyme–dependent RNA modifications. For example, one condition could be DRS data from wild-type human cells paired with DRS data from a sample where METTL3, an m6A methyltransferase, is knocked down (Leger et al., 2021). Given this experimental design, Nanocompore can determine the position of METTL3-dependent RNA modifications. Additionally, in theory, it should be possible to detect the enzyme-dependent RNA modifications by overexpressing a modification-writing enzyme. However, to date, this has not been tested directly.
Alternatively, an in vitro –transcribed (IVT) RNA sample can be used as a reference sample (Tavakoli et al., 2022; Zhang et al., 2021). The IVT is completely composed of canonical RNA nucleotides, with no modifications and can be generated from a cDNA template that was produced using the RNA isolated from the target cells (Tavakoli et al., 2022; Zhang et al., 2021). When this IVT sample is compared to the native RNA sample with Nanocompore, the positions of all RNA modifications are detected, but not their identity. Conceivably, this permits detecting the position of previously unknown RNA modifications, but would require validation with orthogonal techniques, such as mass spectrometry, to characterize and identify them.
Nanocompore can be run with only a single replicate per condition, but this is not advisable. There is no limit to the number of replicates per condition, but it is recommended to use two or three replicates per condition. These replicates are used to filter low-coverage transcripts and add more power to the statistical tests. By default, Nanocompore requires at least 30 reads per position from each replicate; otherwise, the position is ignored. This is a parameter that can be changed to include transcripts with fewer reads if they must be analyzed by Nanocompore. In the current implementation of Nanocompore, the data from each replicate is pooled for the comparative statistical tests. However, using replicates to control for biological and technical variation is part of the future development of Nanocompore.
Additionally, Nanocompore expects the reads to be aligned to a reference transcriptome rather than the genome reference. This eliminates the need for splicing-aware alignments, which can introduce inaccuracies, and permits detecting differential RNA modifications of transcript isoforms. Furthermore, this choice improved the analysis parallelization and improved the protocol run time. Many organisms have well established reference transcriptome files, such as humans (Frankish et al., 2021), yeast (Cherry et al., 2012), or mouse (Frankish et al., 2021). However, if the organism of interest does not have a readily available reference transcriptome, one must be assembled from the data before analysis with Nanocompore. Transcriptome assembly is outside the scope of this protocol, and we refer interested readers to the extensive literature on the subject (Shumate, Wong, Pertea, & Pertea, 2022; Tang et al., 2020) .
An important consideration before undertaking this approach is that nanopore data can take up a large amount of storage space, and certain steps are computationally demanding. It is advisable to use high-performance computation (HPC) clusters or a workstation with at least 60 gigabytes of memory and 3 to 10 terabytes of scratch storage space, depending on the number of replicates and depth of sequencing. Certain steps such as basecalling and signal-to-sequence alignment can be accelerated using a graphical processing unit (GPU), although this is not a requirement.
Basic Protocol: RNA MODIFICATION DETECTION USING NANOCOMPORE
Here, users will start from raw nanopore sequencing data for use in Nanocompore for a comparative statistical test to identify RNA modifications between two conditions, a test and a reference condition. The raw fast5 files will be basecalled into fastq files that will be aligned to a reference transcriptome. Using the aligned bam file, the fastq files, and the fast5 files, the ionic current for each read in the fast5 files will be aligned to the reference using a signal-to-sequence alignment algorithm (resquiggling). This provides a mapping of all the ionic current features from each read to specific positions in the reference transcriptome. These signal-to-sequence alignment files can divide the ionic current data for a single position into multiple lines, and thus these files need to be preprocessed for the position-by-position statistical comparison between experimental conditions to detect RNA modifications. If executed correctly, there will be three database files, a log file, and a nanocompore_results.tsv file that details the probability of detecting a modification at each position in the reference file with sufficient coverage. This workflow is also described online at https://nanocompore.rna.rocks/.
Materials
Hardware
Computational environment with at least 3 cores
Software
- python (version 3.7) (Python Language Reference 2009)
- ONT guppy basecaller (version 4.4.2) (https://community.nanoporetech.com)
- Guppy version 4.4.2 was used in developing and testing Nanocompore. Newer versions of the guppy basecaller are available from Oxford Nanopore Technologies and appear to work as expected with Nanocompore.
- minimap2 (version 2.20-r1061) (https://github.com/lh3/minimap2) (Li, 2018)
- samtools (version 1.12) (http://www.htslib.org) (Danecek et al., 2021)
- nanopolish (version 0.13.3) (https://github.com/jts/nanopolish) (Simpson et al., 2017)
- nanocompore (version 1.0.3) (https://nanocompore.rna.rocks) (Leger et al., 2021)
- Nanocompore can be installed with at least three different methods. See https://nanocompore.rna.rocks/installation/for detailed up-to-date installation instructions.
Files
- Fast5 file(s) from experimental condition 1 (typically, these are wild-type samples where RNA modifications are to be identified)
- Fast5 file(s) from experimental condition 2 (typically, these are a KO/KD of a particular modification writing enzyme which are greatly reduced or devoid of the modification of interest)
- _The data availability section has links to the data used for this manuscript and they can be downloaded from their cited repositories. A detailed description of the fast5 format can be found in supplementary note 1 from the creators of the SLOW5 format (_Gamaarachchi et al. , 2022 ).
- Reference transcriptome fasta file [e.g., gencode.v37.transcripts.fa (Frankish et al., 2021)]
CAUTION : Different fonts can change some special characters, e.g., the hyphen; therefore you may need to write out the commands instead of copy-pasting them as directed in this protocol.
CAUTION : All installed software are assumed to be in the bash path.
CAUTION : This protocol has been tested on the listed software versions. Alternate software versions may still function as expected.
CAUTION : For the commands below, the files are assumed to be in the directory from where the command is executed. A file path must be provided for each file that is in a different directory.
1.Basecall the Fast5 files with the latest version of the guppy basecaller:
- guppy_basecaller -i {raw_fast5_dir} -s {dest_dir} --flowcell {flowcell_id} --
- kit {kit_id} -r --calib_detect --enable_trimming true --trim_strategy rna --
- reverse_sequence true
2.Concatenate the output fastq files into a single fastq file:
- cat {dir_to_guppy_output}/*.fastq > {basecalled_fastq}
3.Align the fastq files to a reference using minimap2 and filter the alignments with samtools:
- minimap2 -ax map-ont -L {transcriptome_fasta} {basecalled_fastq} | samtools
- view -bh -F 2324 -q 10 | samtools sort -O bam > {aligned_reads_bam}
4.Index the sorted and filtered bam file with samtools:
- samtools index {aligned_reads_bam}
5.Index the fastq file with the corresponding fast5 files using nanopolish:
- nanopolish index -s {sequencing_summary.txt} -d {raw_fast5_dir}
- {basecalled_fastq}
6.Align the ionic current segments to the nucleotide sequence with nanopolish:
- nanopolish eventalign --reads {basecalled_fastq} --bam {aligned_reads_bam} --
- genome {transcriptome_fasta} --print-read-names --scale-events --samples >
- {eventalign_reads_tsv}
7.Collapse the eventalign_reads_tsv file into unique kmers per read with Nanocompore eventalign_collapse:
- nanocompore eventalign_collapse -t {n_threads} -i {eventalign_reads_tsv} -o
- {eventalign_collapsed_reads_tsv}
8.Repeat steps 1 through 7 for each sequencing experiment for both conditions.
9.Compare the ionic current characteristics of reads from two conditions to detect the presence of RNA modifications with Nanocompore sampcomp:
- nanocompore sampcomp -1 {C1_1},{C1_2},...{C1_N} -2 {C2_1},{C2_2},...{C2_N} -f
- {transcriptome_fasta} --label1 {Cond1} --label2 {Cond2} -o
- {dir_output_results}
Alternate Protocol 1: RNA MODIFICATION DETECTION BY NANOCOMPORE WITH f5c
This alternate protocol is conceptually similar to the Basic Protocol and will follow many of the same steps: converting the ionic current signals into nucleotides (basecalling); aligning the resulting fastq file to the reference transcriptome file; collapsing the resquiggled data; and conducting the positional pairwise comparison between sample conditions. However, the alignment of the ionic current segments to the reference transcriptome (resquiggling) is done using the software f5c (Gamaarachchi et al., 2020) instead of nanopolish. F5c implements the same functionality of nanopolish but has a significantly faster runtime. At the time of Nanocompore development, f5c was not yet available, so the official workflow presented by Leger et al. (2021) uses Nanopolish. For consistency, our default protocol also uses Nanopolish; however, users are encouraged to apply this alternate protocol instead, which leads to identical results with lower execution time. The output files from this alternate protocol are identical to those from the Basic Protocol, namely three database files, a log file, and a nanocompore_results.tsv file that contains the probability of containing a modification for each position in the reference with sufficient coverage (by default 30 reads per replicate). The p -values from the statistical tests in the nanocompore_results.tsv file can be filtered based on the users’ thresholds to determine the positions of the modifications of interest.
Additional Materials (also see Basic Protocol)
Software
f5c (version 0.6) (https://github.com/hasindu2008/f5c) (Gamaarachchi et al., 2020)
1.Follow steps 1-4 of the Basic Protocol.
2.Index the fastq file with the corresponding fast5 files with f5c:
- f5c index --iop {Number of Input/output processes} -s {sequencing_summary.txt}
- -d {raw_fast5_dir} {basecalled_fastq}
3.Align the ionic current segments to the nucleotide sequence with f5c:
- f5c eventalign -t {Number of processing threads} --iop {Number of Input/output
- processes} -r {basecalled_fastq} -b {aligned_reads_bam} -g
- {transcriptome_fasta} --rna --secondary=yes, --min-mapq 0 --print-read-names -
- -scale-events --samples -o {eventalign_reads_tsv}
4.Collapse the eventalign_reads_tsv file into unique kmers per read with Nanocompore eventalign_collapse:
- nanocompore eventalign_collapse -t {n_threads} -i {eventalign_reads_tsv} -o
- {eventalign_collapsed_reads_tsv}
5.Repeat steps 1 through 4 from Alternate Protocol 1 for each sequencing experiment for both conditions.
6.Follow step 9 of the Basic Protocol.
Alternate Protocol 2: RNA MODIFICATION DETECTION BY NANOCOMPORE USING NEXTFLOW
This alternate protocol details how to run the entire Nanocompore pipeline using a nextflow pipeline manager. This has the advantage that all the dependent software tools (such as guppy, samtools, f5c, and Nanocompore) are contained within images, which improves the reproducibility of the protocol by limiting software installation and system variation. The user will create a sample.txt file which contains the sample labels, condition labels, and paths to the corresponding directory of fast5 files, which will allow the pipeline to access the correct data. Then, the nextflow pipeline must be configured with paths to files such as the references, the Nanocompore parameters, and the output directory path. The final step is to initiate the nextflow pipeline, which will then execute all the steps detailed in the Basic Protocol. Output from this alternate protocol is identical to that from the Basic Protocol (three database files, a log file, and a nanocompore_results.tsv file). The nanocompore_results.tsv file contains the probability of a modification existing at each position in the reference, which can be filtered based on users’ thresholds to determine the modified positions.
Materials
Hardware
Computational environment with at least 3 cores
Software
- nextflow (version 22.04.5.5708) (Di Tommaso et al., 2017)
- singularity (version 3.5.3-1.1.el8) (Kurtzer, Sochat, & Bauer, 2017)
- nanocompore nextflow pipeline (https://github.com/tleonardi/nanocompore_pipeline)
Files
- Fast5 file(s) from experimental condition 1 (typically, these are wild-type samples where RNA modifications are to be identified)
- Fast5 file(s) from experimental condition 2 (typically, these are a KO/KD of a particular modification-writing enzyme which are greatly reduced or devoid of the modification of interest)
- _The data availability section has links to the data used for this manuscript, and they can be downloaded from their cited repositories. A detailed description of the fast5 format can be found in supplementary note 1 from the creators of the SLOW5 format (_Gamaarachchi et al. , 2022 ).
- Reference transcriptome fasta file (e.g., gencode.v37.transcripts.fa (Frankish et al., 2021))
1.Prepare the tab separated sample.txt file with the following format.
| SampleName | Condition | DataPath |
| KD1 | KD | /path/to/KD1_fast5_directory |
| KD2 | KD | /path/to/KD2_fast5_directory |
| WT1 | WT | /path/to/WT1_fast5_directory |
| WT2 | WT | /path/to/WT2_fast5_directory |
2.Configure the nextflow pipeline by editing the nextflow.config.template file
3.Run the nextflow pipeline:
- nextflow run pipeline.nf
COMMENTARY
Background Information
Nanopore sequencing became commercially available from Oxford Nanopore Technologies (ONT) in 2014, and the direct RNA nanopore sequencing protocol was released in 2016. A major strength of DRS is that RNA modifications are kept intact during sequencing, and interact with the nanopore in different ways than the canonical counterparts. These RNA modifications can change the raw signal in many ways, typically by altering the ionic current intensity or the amount of time the motor enzyme takes to translocate the next nucleotide through the pore (dwell time). Detecting these RNA modifications became a priority for the field once DRS became available, with at least fifteen software tools developed within the first four years of this technology being released to the public (Furlan et al., 2021). The first attempts used systematic basecalling and alignment errors to detect the presence of modifications in the RNA strands (Abebe et al., 2022; Begik et al., 2021; Jenjaroenpun et al., 2021; Smith et al., 2019). Following this, tools were developed that used the raw ionic current to detect RNA modifications instead of relying on systematic errors from the ONT basecalling algorithm (Begik et al., 2021; Gao et al., 2021; Leger et al., 2021; Lorenz et al., 2020; Pratanwanich et al., 2021).
Due to the complexity of generating training data for modification detection, we developed Nanocompore (Leger et al., 2021), which took the alternative route of using an unsupervised nonparametric algorithm, thus not requiring model training. The rationale behind Nanocompore is based on the observation that modified nucleotides have a characteristic effect on the ionic current and dwell time relative to their unmodified counterparts. This observation was first exploited by the Tombo algorithm (Stoiber et al., 2017), developed by ONT, to test for differences in the electrical signal between a sample of interest and an unmodified reference. However, this approach proved to have a comparatively high false positive rate, which severely limited its applicability. Building on this work, Nanocompore introduced a clustering step with Gaussian Mixture Modeling (GMM) to identify two distinct nucleotide clusters in the bivariate space formed by current intensity and dwell time. Following the identification of the two clusters, Nanocompore applies one of multiple statistical tests to assess whether the reads are distributed differently among the two clusters in the sample of interest relative to the reference, unmodified sample. In the original work that introduced the Nanocompore algorithm, the authors performed several benchmarks, both in silico and in vitro , to measure its sensitivity and specificity (Leger et al., 2021). These benchmarks revealed that, compared to most other methods available, Nanocompore achieves high specificity in the detection of m6A.
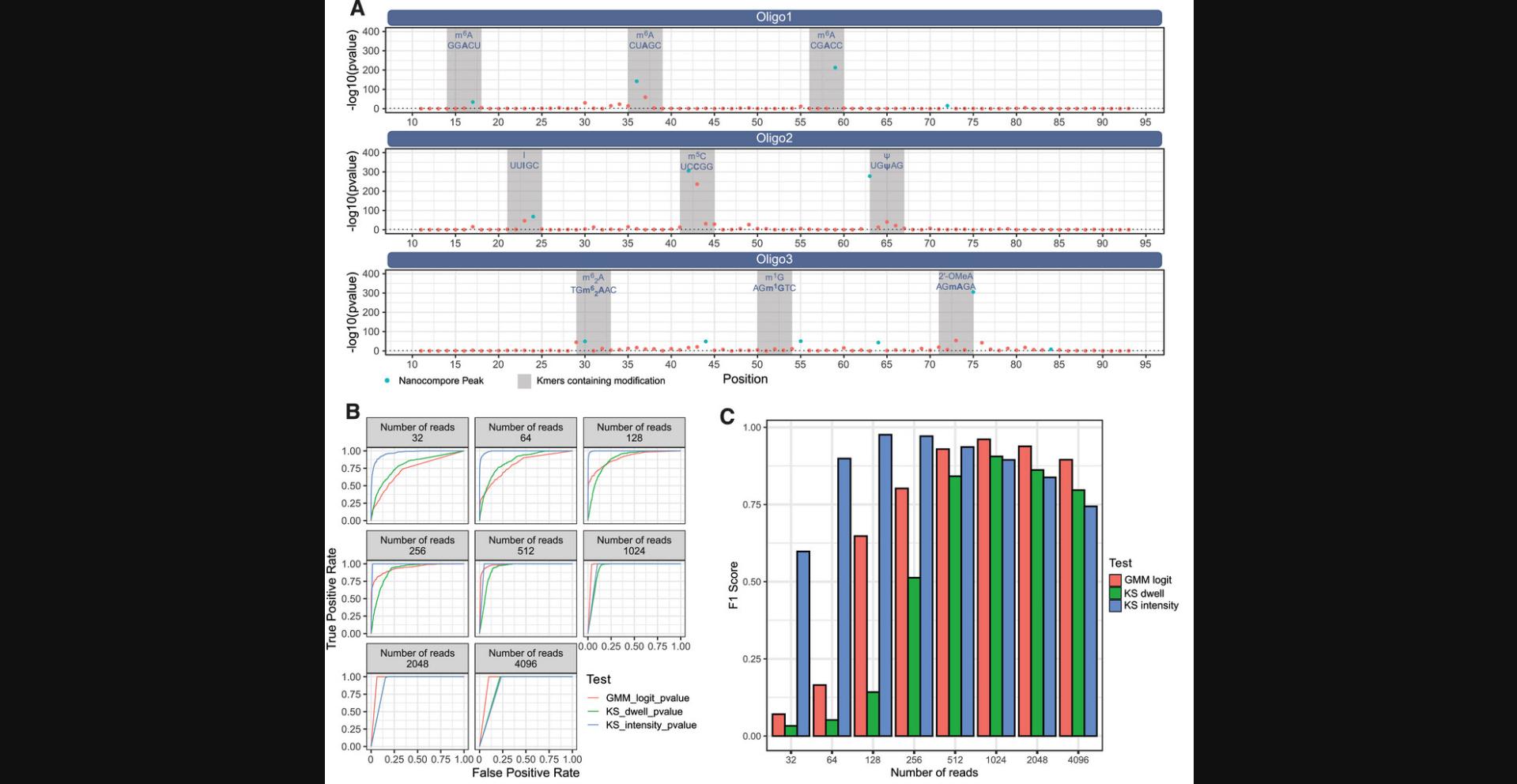
The authors were also able to show that Nanocompore can be used to detect other modifications in addition to m6A. Specifically, the authors used four oligonucleotides each containing three RNA modifications at specific sites (m6A, I, m5C, Ψ, m6,2A, m1G and 2′-OMeA) and the fourth was devoid of any RNA modifications (Leger et al., 2021). These oligonucleotides were processed with the Nanocompore protocol using the unmodified oligonucleotide as the reference sample, and then one of the three modified oligonucleotides as the test sample. The most significant p -values detected for each modified nucleotide were located within the window of positions that can be affected by a single modified nucleotide (Fig. 2A). To evaluate how the three default statistical tests performed with varying levels of coverage, the reads from oligonucleotide 1 (containing three m6A modified positions) and the unmodified oligonucleotide were randomly downsampled to eight different read counts (32, 64, 128, 256, 512, 1024, 2048, and 4096) and compared with the Nanocompore protocol (Fig. 2B). The false positive and true positive rates for each of the three tests at each coverage depth were determined for identifying the three m6A sites in oligonucleotide 1, and demonstrate that there is a major detrimental impact on detection accuracy, as sequencing depth is reduced. Furthermore, an F1 score (a statistic to test a model's accuracy, which is the harmonic mean of precision and recall) was also computed for each condition to visualize the effect of coverage on correctly identifying m6A from oligonucleotide 1 (Fig. 2C). This benchmarking demonstrates that Nanocompore can correctly identify m6A with more than 30 reads, but ideally more than 100 reads per sample.
The authors of Nanocompore compared five other RNA modification detection tools to Nanocompore on a biological dataset (Leger et al., 2021). The yeast METTL3 homolog is IME4, and poly(A) RNA from wild-type sk1 yeast and an IME4 knockout strain were sequenced using DRS. The six tools were used to detect m6A, and Nanocompore was found to have the highest specificity for detecting m6A (Leger et al., 2021). Nanocompore's high specificity was confirmed with synthetic IVT oligonucleotides where all adenosines were replaced by m6A or all the cytosines were replaced by m5C, and Nanocompore found no false positive sites (Acera Mateos et al., 2022).
Additionally, Nanocompore compares two conditions, and as such, alterations to the nanopore sequencing platform are controlled by comparing the RNA samples instead of just modeling ionic current features of particular RNA modifications. This is important, because changes such as buffer composition, motor enzyme, or the nanopore, can all affect the ionic current measured by the sequencer. Furthermore, Nanocompore can, in principle, detect all RNA modifications, as long as the reference sample is chosen wisely. Other software tools that detect RNA modifications have high accuracy only for specific RNA modification types, or are limited to particular versions of the nanopore platform (Furlan et al., 2021). Non-nanopore-based RNA modification detection techniques, such as mass spectrometry, miCLIP, and primer extension experiments, are limited to a single or few RNA modification types or limited by the quality of the biophysical assay to detect RNA modifications, or lack sequence specificity (Helm & Motorin, 2017). Hence DRS can be used to determine the prevalence of multiple RNA modifications on the same reference transcript, or even each single molecule sequenced (Furlan et al., 2021).
Mapping RNA modifications in the transcriptome can be used to determine the functional consequences of certain RNA modifications (Anreiter et al., 2021). RNA modifications are known to be important for proper transcript maturation, function, and eventual decay (Barbieri & Kouzarides, 2020). Alterations to RNA modifications can result in dysfunctional transcripts and lead to disease (Barbieri & Kouzarides, 2020). Coupling RNA modification detection with long-read sequencing can be used to evaluate the role of RNA modifications on particular isoforms (Workman et al., 2019) or determine if certain modifications are independent or dependent on each other (Leger et al., 2021).
Critical Parameters
Nanocompore compares two samples using statistical testing to identify RNA modifications. These statistical tests are more accurate with higher numbers of reads per position. Due to this, by default, Nanocompore requires a minimum of 30 reads per replicate for each position. This threshold can be changed by the user to enforce more stringent or more relaxed coverage requirements. For certain highly expressed transcripts, this coverage requirement is usually exceeded. However, transcripts with lower expression may have positions skipped that are of interest because one or more DRS samples fall below this coverage threshold. It is not advisable to reduce the coverage threshold, because the tests become more unreliable and prone to outlier effects, but it can be reduced if a critical low expression transcript must be analyzed by Nanocompore. Alternatively, biochemistry techniques such as biotinylated probes can enrich a target transcript or use software such as readuntil to deplete undesired transcripts during sequencing (Weilguny et al., 2023).
Additionally, by default, the minimum reference length is 100 nt long. This is set to 100 nt because many of the data processing steps are less reliable on shorter transcripts. The basecaller requires some sequence to anchor the signal and begin basecalling, the long-read mapping algorithms align shorter reads less reliably, and the resquiggling process implemented by nanopolish has a hard-coded minimum read length threshold of 100 nt. The default length threshold in Nanocompore can be changed to shorter than 100 if the target gene of interest is shorter than 100 nt, but users would also need to ensure that this length threshold is also changed in nanopolish and all other data processing steps.
Troubleshooting
See Table 1 for a list of common problems with the protocols, their causes, and potential solutions.
| Problem | Possible cause | Solution |
|---|---|---|
| Very few or no significant p values reported | Modification reduction from reference sample was incomplete | Check knockdown efficiency, and possibly redo knockdown. It is also always advisable to include positive controls that are known to be modified. |
| High number of kmers not analyzed | One sample has coverage lower than the “lower limit” threshold (default coverage threshold of 30 reads per transcript) | Remove the sample or concatenate that sample with another from the same experimental condition |
| Nanocompore exits with an error | One or more files prepared for Nanocompore sampcomp have errors | Check Nanocompore log files for the file, kmer, or gene causing the error and correct the pre-processing error that caused that error |
| GMM p-value is "NC" | The GMM algorithm could not identify two clusters for this position; hence the position was skipped from p-value calculation. There are multiple reasons underlying this problem, for example noisy data or insufficient coverage. | If several positions are marked as NC, increasing the coverage could improve the likelihood of the GMM algorithm correctly identifying two clusters |
| Nanocompore terminates immediately, producing an empty results file | The most common cause for this problem is that all transcripts were discarded during the whitelisting step | Try running Nanocompore lowering the minimum coverage threshold. If the generated results file contains transcript, then the analysis needs to be repeated with higher coverage, e.g., by sequencing additional replicates. |
| The results file does not report the genomic coordinate of the sites identified | Nanocompore was run without a BED annotation | Run Nanocompore again passing the transcriptome annotation in BED format as an input |
Understanding Results
Nanocompore result table
Nanocompore sampcomp will produce a table of statistical test results for each kmer with sufficient coverage (by default 30 reads per replicate) in both experimental conditions (Table 2). The table has basic information about each kmer, such as the transcriptomic position, the reference name, and the reference kmer sequence. Importantly, the transcriptomic position corresponds to the first nucleotide of the reported kmer. Additionally, if the optional bed file of genomic coordinates for each gene was provided, the genomic coordinates will also be included in the table. The genomic coordinates will be represented with “NA” if the optional bed file was not provided. For each of the kmers, the table reports the multiple testing–corrected p -values for each statistical test that Nanocompore conducts. By default, this includes: the p -value from the Kolmogorov-Smirnov (KS) test of ionic current intensities, the p -value from the KS test of kmer dwell time, the GMM p -value, the type of GMM cluster used, the number of GMM clusters, the number of reads from each sample assigned to each cluster, and the log odds ratio (LOR) of sample labels assigned to each cluster.
| pos | chr | genomicPos | ref_id | strand | ref_kmer | GMM_logit_p-value | KS_dwell_p-value | KS_intensity_p-value | GMM_cov_type | GMM_n_clusters | cluster_counts | Logit_LOR |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 11 | NA | NA | control | NA | AGAUA | 0.586350 | 0.302508 | 0.525576 | Full | 2 | oligo1_1:44/412__control_1:45/489 | 0.148978 |
| 12 | NA | NA | control | NA | GAUAG | 0.499759 | 0.701376 | 0.001443 | Full | 2 | oligo1_1:636/40__control_1:832/65 | 0.207818 |
| 13 | NA | NA | control | NA | AUAGG | 0.352552 | 0.853331 | 0.899235 | Full | 2 | oligo1_1:1138/25__control_1:1621/50 | 0.320219 |
| 14 | NA | NA | control | NA | UAGGA | 0.452006 | 0.138293 | 9.40e-08 | Full | 2 | oligo1_1:1008/302__control_1:1507/411 | -0.094533 |
| 15 | NA | NA | control | NA | AGGAC | 0.013222 | 2.21e-08 | 0.003626 | Full | 2 | oligo1_1:1179/400__control_1:1590/670 | 0.215959 |
| 16 | NA | NA | control | NA | GGACU | 8.11e-17 | 1.47e-14 | 1.47e-14 | Full | 2 | oligo1_1:2120/126__control_1:3004/446 | 0.909981 |
| 17 | NA | NA | control | NA | GACUC | 0.000321 | 4.97e-07 | 3.46e-14 | Full | 2 | oligo1_1:2438/20__control_1:3580/83 | 1.002240 |
| 18 | NA | NA | control | NA | ACUCU | 0.002309 | 0.152668 | 0.004484 | Full | 2 | oligo1_1:2344/129__control_1:3396/276 | 0.385875 |
| 19 | NA | NA | control | NA | CUCUU | 0.260900 | 0.853331 | 0.017137 | Full | 2 | oligo1_1:92/2427__control_1:164/3507 | -0.205367 |
| 20 | NA | NA | control | NA | UCUUU | 0.002555 | 0.506629 | 0.323974 | Full | 2 | oligo1_1:2745/111__control_1:3551/218 | 0.413207 |
- a The pos column refers to the reference transcript nucleotide position. The chr column refers to the scaffold name of the genomic coordinate. The genomic Pos column refers to the nucleotide position in the genome. Chr and genomic Pos will be NA unless a bed file is provided that maps the transcriptome coordinates to the genome coordinates. The ref_id column refers to the transcript reference name. The strand column refers to which of the two reference chromosomes the transcript is found on. This will be NA unless a bed file is provided that maps the transcriptome coordinates to the genome coordinates. The ref_kmer column is the sequence identity of the reference kmer. The GMM_logit_p-value column refers to the p-value produced by the GMM clustering and logistical regression statistical test for that particular kmer. The KS_dwell_p-value and KS_intensity_p-value columns refer to the p-value produced by the Kolmogorov–Smirnov test on the dwell time and ionic current intensity distributions respectively. The GMM_cov_type column refers to the initial shape of the GMM clustering. The GMM_n_clusters column refers to the number of clusters, 1 or 2, that best fit the data. The clusters_counts column details the number of reads that were assigned to each cluster from each experimental condition. The Logit_LOR column refers to the logistical regression odds ratio (LOR). The absolute value of the LOR is what is useful for filtering significant GMM p-value. All columns that are derived from the GMM logit test will be NA if only one cluster best represents the data instead of two.
- Numerical values have been reduced to 6 decimal places for display in the table.
The KS test is a non-parametric test that is used to determine if two sample distributions are derived from the same underlying population. A significant p -value means that the distribution of values for a particular kmer is different between the two conditions. The KS test is performed separately on the median ionic current and the dwell time for each kmer. A significant p -value from either test can indicate that a modification is present.
The GMM clusters all the data for a kmer from both samples in two dimensions—median ionic current and dwell time using either one or two clusters. No further testing is required, and the kmer is called unmodified if only one cluster fits the data better than two clusters. However, if two clusters fit the data better than one cluster, the sample labels are assigned to one of the two corresponding clusters, and a logistical regression (logit) test is performed on the sample label assignments. A significant p -value means that a sample label is enriched for one of the two clusters, indicating that a modification is likely present. The LOR value is generated in addition to a p -value from the GMM Logit test. The LOR value can be used in conjunction with the GMM p -value to filter for the presence of higher-probability modifications. The absolute value of LOR 0.5 means that the probability of a read from one condition is ∼3 times more likely in one cluster than a read from the other condition. A larger absolute value of LOR means that reads from one condition are more heavily enriched in one cluster, and an absolute value of LOR closer to 0 means that the two sample labels are more evenly distributed between the two clusters.
Interpreting the values in the results table
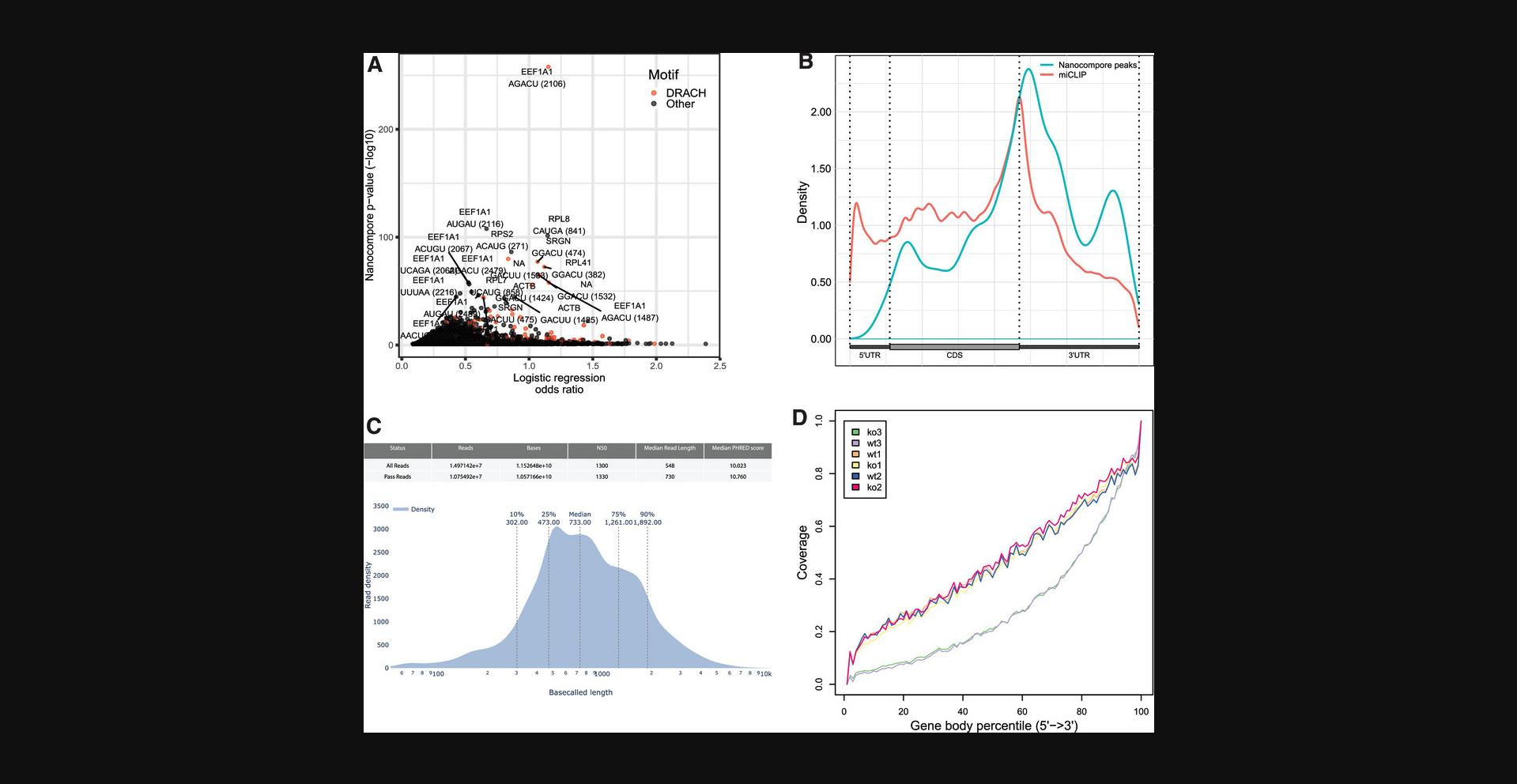
Typically, the GMM p -value ≤ 0.01 [or –log10(GMM p -value)≥2] and the absolute value of the LOR > 0.5 are used together to determine if a kmer is likely modified. Plotting the –log10 of the GMM p -value on the y-axis and the absolute value of the LOR on the x-axis is a method to quickly visualize how many significant kmers were identified by Nanocompore (Fig. 3A). What tends to become apparent is the number of sites that have a significant GMM p -value but have a LOR < 0.5. Using LOR as an additional threshold for determining modified sites improves specificity but lowers sensitivity. Alternatively, the KS test on the individual components of the ionic current (intensity and dwell time) can be used to determine the presence of an RNA modification. The KS test on the ionic current intensity outperformed the GMM when read coverage was low on synthetic RNA oligonucleotides where the modification sites were known and at ≥99% modified (Fig. 2C). Although the GMM test will generally outperform the KS test on biological data (Leger et al., 2021), it is important to keep the results of the KS test in mind when evaluating low-coverage positions.
Interpreting neighboring significant positions
Regardless of which test was used, a single modified nucleotide can cause ionic current changes in more than one position. The sensitive region of the CsgG nanopore within version 9 MinION flowcells contacts ∼5 RNA nucleotides simultaneously. This means that a single nucleotide exists within 5 kmers as it translocates through the nanopore. A nucleotide typically has the maximal impact on the ionic current when it is at the center of the kmer and affects the ionic current less the further from the kmer center. However, kmer context can alter which position the RNA modification can alter compared to the unmodified kmer. Some kmers have similar ionic current intensities and the high variance can sometimes obscure the influence of an RNA modification on the ionic current signal. This means that the modification-dependent ionic current changes can be detected in one or more kmers that contain it, depending on the surrounding context and noise profile of those kmers. This is something that should be considered when interpreting adjacent kmers that are significant for a modification. One method to reduce the effects of one modification altering multiple kmers is to use a peak-calling algorithm on the –log10 of the p -value. This will filter for the most significant p -values that are within a certain distance of each other. Typically, peak calling with a window size of 5 and a floor of 2 will reduce this effect.
Alternatively, adjacent significant p -values could indicate that there are multiple adjacent modifications as documented in rRNA and tRNA molecules (Suzuki, 2021; Taoka et al., 2018). Identical adjacent RNA modifications will confound each other, and Nanocompore cannot resolve them. However, different adjacent modifications can be controlled for, depending on the reference sample.
Importance of the reference sample
The most important consideration for interpreting Nanocompore p -values is the reference sample that was used for the comparison. The simplest experimental design to interpret is when a single modification-writing enzyme is knocked out of one sample and the other sample is a wild-type version of the same cells. This design controls for things like single-nucleotide polymorphisms, multiple types of modifications obscuring results, different types of modifications near each other, and other confounding variables (Leger et al., 2021). A significant p -value can confidently be assigned as an RNA modification dependent on the knocked-out enzyme. An experimental design that requires more consideration is when an IVT RNA sample from the same genetic background as the modified sample is used. This is because the positions of all RNA modifications can be detected simultaneously; however, the identity of the RNA modification is not determined. Additionally, adjacent RNA modifications are even more challenging to resolve because no RNA modification remains the same between the two conditions. This effect is controlled for when using a modification-writing enzyme knockout or knock down when the adjacent modification is not dependent on the knocked-out or knocked-down enzyme because it will still be present in both samples. When using an IVT reference, further information is required to determine the identity of modifications detected. Significant kmers that overlap with positions of known modifications or sequence motifs that are typically modified by certain enzymes can be used to infer the modification identity. Follow-up experiments using short read sequencing that targets certain modifications, biophysical assays [such as thin-layer chromatography (Keith, 1995)], mass spectrometry (Cao & Limbach, 2015), or DRS where a target enzyme is knocked down or knocked out [such as a defective vir-1 in Arabidopsis (Parker et al., 2020) or METTL3 knockout in humans (Leger et al., 2021)] can determine the identity of modifications detected this way. However, an IVT reference sample has the potential to discover the position of novel RNA modifications, which other target-specific techniques cannot accomplish, because it makes no assumptions about the identity of the RNA modification. Any site that is suspected to be a novel modification should be further characterized with mass spectrometry or biophysical assays (such as two-dimensional gels) that target the transcript or position containing the putative novel modification.
The most difficult experimental design to consider is when the genetic backgrounds of the two samples are different. This is because SNPs can cause kmers with different nucleotide compositions to be aligned to the same reference position despite having different sequences. In much the same was as an RNA modification causes alterations to the ionic current relative to the unmodified form, two different nucleotides will have different ionic currents relative to each other (Stoddart, Heron, Mikhailova, Maglia, & Bayley, 2009). In this case, Nanocompore could detect an ionic current difference and report a significant p -value because Nanocompore cannot discriminate between a SNP and an RNA modification if there is a difference in raw ionic current for the comparison. One method to overcome SNPs and other genetic differences is to use DNA sequencing or short-read RNA-seq and variant calling to identify all positions that are due to genetic differences and not due to RNA modification differences. Blacklisting positions or regions limits the scope of RNA modification detection, but reduces the number of incorrectly identified RNA modification sites.
Quality Control
Input data quality control
There are several ways to evaluate that the analysis performed by Nanocompore was completed correctly. The first of these is to ensure that the DRS data used by Nanocompore is of sufficient quality. The data from DRS experiments can vary from run to run, depending on features like the quality of the input RNA or the number of nanopores in the flowcell. The software package pycoQC plots many of the DRS QC metrics in an interactive web format (Leger & Leonardi, 2019). Two significant features that pycoQC plots are the number of reads or Gigabases (GB) sequenced and the read length distribution. A typical MinION flowcell is expected to produce between 1.5 and 2.5 million reads that have a mean read length of 700 nt or a read N50 of 1350 nt from human poly(A) RNA (Fig. 3C). The read N50 is the read length where half of the total sequenced nucleotides are contained in reads of this length or longer. The read N50 is a good metric for nanopore performance because shorter reads are sequenced faster than longer reads, and although the majority of nucleotides sequenced may be in much longer reads, the number of shorter reads skews the mean in a shorter direction. Significantly fewer reads or shorter mean or read N50 lengths may indicate that there was an issue with the input RNA or the MinION flowcell, and modification detection analysis with Nanocompore may not have enough coverage to detect RNA modifications at some positions.
In addition to the metrics from pycoQC, average coverage across the transcriptome can be informative as to the quality of the DRS data. Calculating the sequencing depth at every transcript position and plotting the normalized coverage across the quantile transcript position for all transcripts can be used to determine the data quality. One method to create this transcriptome-wide, normalized, quantile plot is with the geneBody_coverage.py script in the RSeQC package (Wang, Wang, & Li, 2012). RNA is read in the 3′-to-5′’ orientation during DRS experiments, and thus it is expected to have more coverage at the 3′ end than the 5′ end due to RNA degradation and sequencing artifacts (Workman et al., 2019). The rate of coverage loss from the 3′ end to 5′ end can be informative about the quality of the input RNA and explain why certain positions may have been skipped during sequencing. For example, two out of the six yeast replicates from Leger et al. (2021) show much lower coverage across the transcriptome than the other four (Fig. 3D). This is likely due to more fragmented RNA used in the sequencing library than the other four replicates, and could indicate that these replicates should be discarded from the analysis, resequenced, or used with care.
Analysis quality control
There are two QC indications that Nanocompore was completed successfully. The first is to review the log file. This file will contain information about each step of Nanocompore and can be used to quickly review which transcripts and positions were skipped for the analysis and the reason. The log file will also contain any error messages if Nanocompore encountered any critical errors that required it to abort. The second is to ensure that the nanocompore_results.tsv file is formatted correctly and that there are non-zero values in the data columns.
One method to assess the modified RNA positions identified by Nanocompore is to use an internal standard of known RNA modifications. Using an internal standard of a known RNA modification will directly assess that Nanocompore successfully identified an RNA modification. This can be done with a spike-in RNA oligonucleotide that contains one or more RNA modifications at known positions. However, long synthetic modified RNA oligonucleotides are expensive reagents. A more scalable alternative is to compare the RNA modifications identified by Nanocompore to published datasets using orthogonal methods to detect the same type of RNA modification. For example, the distribution of m6A in the transcriptome has been characterized by many methods [such as miCLIP (Linder et al., 2015) or MAZTER-seq (Garcia-Campos et al., 2019)] and is known to be concentrated near mRNA stop codons. The m6A sites identified by Nanocompore can be compared to the m6A sites identified by miCLIP to evaluate the reliability of m6A sites identified by Nanocompore (Fig. 3B; Leger et al., 2021).
Time Considerations
Depending on available computational resources and sequencing depth, the entire protocol generally takes between 1 day and 2 weeks to complete. There are two data processing steps that are the bottleneck of the pipeline: the signal-to-sequence alignment (nanopolish eventalign) step; and formatting the output of the signal-to-sequence alignments into a Nanocompore readable format (Nanocompore eventalign_collapse). However, once these steps are complete, the eventalign_collapsed_reads_tsv files can be reused for any future comparisons, saving time for follow-up analysis.
Acknowledgments
L.M. is supported by the EMBL ETPOD fellowship. F.N. is supported by grants from the Associazione Italiana per la Ricerca sul Cancro (AIRC - IG22851) and from the Next Generation EU (National Center for Gene Therapy and Drugs based on RNA Technology).
Open Access Funding provided by Istituto Italiano di Tecnologia within the CRUI-CARE Agreement.
Author Contributions
Logan Mulroney : writing original draft, writing review and editing; Ewan Birney : supervision, writing review and editing; Tommaso Leonardi : supervision, writing review and editing; Francesco Nicassio : supervision, writing review and editing.
Conflict of Interest
L.M. and T.L have received financial support from ONT for travel and accommodations to attend and present at ONT events. E.B. is a paid consultant and shareholder of ONT. F.N. declares no conflicts of interest.
Open Research
Data Availability Statement
The data used in this protocol (except for the pycoQC plot in Fig. 3) are from Leger et al. (2021) and can be found at the European Nucleotide Archive database under accession codes PRJEB44511 and PRJEB35148. The data used to make the pycoQC plot are from Workman et al. (2019) and can be found at the github repository https://github.com/nanopore-wgs-consortium/NA12878/blob/master/RNA.md.
Literature Cited
- Abebe, J. S., Price, A. M., Hayer, K. E., Mohr, I., Weitzman, M. D., Wilson, A. C., & Depledge, D. P. (2022). DRUMMER—rapid detection of RNA modifications through comparative nanopore sequencing. Bioinformatics , 38(11), 3113–3115. doi: 10.1093/bioinformatics/btac274
- Acera Mateos, P., Sethi, A. J., Ravindran, A., Guarnacci, M., Srivastava, A., Xu, J., … Eyras, E. (2022). Simultaneous identification of m6A and m5C reveals coordinated RNA modification at single-molecule resolution. bioRxiv , doi: 10.1101/2022.03.14.484124
- Anreiter, I., Mir, Q., Simpson, J. T., Janga, S. C., & Soller, M. (2021). New twists in detecting mRNA modification dynamics. Trends in Biotechnology , 39(1), 72–89. doi: 10.1016/j.tibtech.2020.06.002
- Barbieri, I., & Kouzarides, T. (2020). Role of RNA modifications in cancer. Nature Reviews Cancer , 20, 303–322. doi: 10.1038/s41568-020-0253-2
- Begik, O., Lucas, M. C., Pryszcz, L. P., Ramirez, J. M., Medina, R., Milenkovic, I., … Novoa, E. M. (2021). Quantitative profiling of pseudouridylation dynamics in native RNAs with nanopore sequencing. Nature Biotechnology , 39(10), 1278–1291. doi: 10.1038/s41587-021-00915-6
- Boccaletto, P., Machnicka, M. A., Purta, E., Piatkowski, P., Baginski, B., Wirecki, T. K., … Bujnicki, J. M. (2018). MODOMICS: A database of RNA modification pathways. 2017 update. Nucleic Acids Research , 46(D1), D303–7. doi: 10.1093/nar/gkx1030
- Boo, S. H., & Kim, Y. K. (2020). The emerging role of RNA modifications in the regulation of mRNA stability. Experimental & Molecular Medicine, 52(3), 400–408.
- Cao, X., & Limbach, P. A. (2015). Enhanced detection of post-transcriptional modifications using a mass-exclusion list strategy for RNA modification mapping by LC-MS/MS. Analytical Chemistry , 87(16), 8433–8440. doi: 10.1021/acs.analchem.5b01826
- Cherry, J. M., Hong, E. L., Amundsen, C., Balakrishnan, R., Binkley, G., Chan, E. T., … Wong, E. D. (2012). Saccharomyces genome database: The genomics resource of budding yeast. Nucleic Acids Research , 40(Database issue), D700–705. doi: 10.1093/nar/gkr1029
- Cui, L., Ma, R., Cai, J., Guo, C., Chen, Z., Yao, L., … Shi, Y. (2022). RNA modifications: Importance in immune cell biology and related diseases. Signal Transduction and Targeted Therapy , 7(1), 334. doi: 10.1038/s41392-022-01175-9
- Danecek, P., Bonfield, J. K., Liddle, J., Marshall, J., Ohan, V., Pollard, M. O., … Li, H. (2021). Twelve years of SAMtools and BCFtools. GigaScience , 10(2), giab008. doi: 10.1093/gigascience/giab008
- Di Tommaso, P., Chatzou, M., Floden, E. W., Barja, P. P., Palumbo, E., & Notredame, C. (2017). Nextflow enables reproducible computational workflows. Nature Biotechnology , 35(4), 316–319. doi: 10.1038/nbt.3820
- Frankish, A., Diekhans, M., Jungreis, I., Lagarde, J., Loveland, J. E., Mudge, J. M., … Flicek, P. (2021). GENCODE 2021. Nucleic Acids Research , 49(D1), D916–23. doi: 10.1093/nar/gkaa1087
- Furlan, M., Delgado-Tejedor, A., Mulroney, L., Pelizzola, M., Novoa, E. M., & Leonardi, T. (2021). Computational methods for RNA modification detection from nanopore direct RNA sequencing data. RNA Biology , 18(sup1), 31–40. doi: 10.1080/15476286.2021.1978215
- Gamaarachchi, H., Lam, C. W., Jayatilaka, G., Samarakoon, H., Simpson, J. T., Smith, M. A., & Parameswaran, S. (2020). GPU accelerated adaptive banded event alignment for rapid comparative nanopore signal analysis. BMC Bioinformatics , 21(1), 343. doi: 10.1186/s12859-020-03697-x
- Gamaarachchi, H., Samarakoon, H., Jenner, S. P., Ferguson, J. M., Amos, T. G., Hammond, J. M., … Deveson, I. W. (2022). Fast nanopore sequencing data analysis with SLOW5. Nature Biotechnology , 40(7), 1026–1029. doi: 10.1038/s41587-021-01147-4
- Gao, Y., Liu, X., Wu, B., Wang, H., Xi, F., Kohnen, M. V., … Gu, L. (2021). Quantitative profiling of N6-methyladenosine at single-base resolution in stem-differentiating xylem of Populus trichocarpa using nanopore direct RNA sequencing. Genome Biology , 22(1), 1–17. doi: 10.1186/s13059-020-02241-7
- Garalde, D. R., Snell, E. A., Jachimowicz, D., Sipos, B., Lloyd, J. H., Bruce, M., … Turner, D. J. (2018). Highly parallel direct RNA sequencing on an array of nanopores. Nature Methods , 15(3), 201–206. doi: 10.1038/nmeth.4577
- Garcia-Campos, M. A., Edelheit, S., Toth, U., Safra, M., Shachar, R., Viukov, S., … Schwartz, S. (2019). Deciphering the ‘mA Code’ via antibody-independent quantitative profiling. Cell , 178(3), 731–747.e16. doi: 10.1016/j.cell.2019.06.013
- Helm, M., & Motorin, Y. (2017). Detecting RNA modifications in the epitranscriptome: Predict and validate. Nature Reviews. Genetics , 18(5), 275–291. doi: 10.1038/nrg.2016.169
- Jenjaroenpun, P., Wongsurawat, T., Wadley, T. D., Wassenaar, T. M., Liu, J., Dai, Q., … Nookaew, I. (2021). Decoding the epitranscriptional landscape from native RNA sequences. Nucleic Acids Research , 49(2), e7. doi: 10.1093/nar/gkaa620
- Keith, G. (1995). Mobilities of modified ribonucleotides on two-dimensional cellulose thin-layer chromatography. Biochimie , 77(1-2), 142–144. doi: 10.1016/0300-9084(96)88118-1
- Kurtzer, G. M., Sochat, V., & Bauer, M. W. (2017). Singularity: Scientific containers for mobility of compute. PloS One , 12(5), e0177459. doi: 10.1371/journal.pone.0177459
- Leger, A., Amaral, P. P., Pandolfini, L., Capitanchik, C., Capraro, F., Miano, V., … Kouzarides, T. (2021). RNA modifications detection by comparative nanopore direct RNA sequencing. Nature Communications , 12(1), 7198. doi: 10.1038/s41467-021-27393-3
- Leger, A., & Leonardi, T. (2019). pycoQC, interactive quality control for Oxford Nanopore sequencing. Journal of Open Source Software , 4(34), 1236. doi: 10.21105/joss.01236
- Li, H. (2018). Minimap2: Pairwise alignment for nucleotide sequences. Bioinformatics , 34(18), 3094–3100. doi: 10.1093/bioinformatics/bty191
- Linder, B., Grozhik, A. V., Olarerin-George, A. O., Meydan, C., Mason, C. E., & Jaffrey, S. R. (2015). Single-nucleotide-resolution mapping of m6A and m6Am throughout the transcriptome. Nature Methods , 12(8), 767–772. doi: 10.1038/nmeth.3453
- Liu, H., Begik, O., Lucas, M. C., Ramirez, J. M., Mason, C. E., Wiener, D., … Novoa, E. M. (2019). Accurate detection of m6A RNA modifications in native RNA sequences. Nature Communications , 10(1), 1–9.
- Liu, H., Begik, O., & Novoa, E. M. (2021). EpiNano: Detection of mA RNA modifications using Oxford Nanopore direct RNA sequencing. Methods in Molecular Biology , 2298, 31–52. doi: 10.1007/978-1-0716-1374-0_3
- Lorenz, D. A., Sathe, S., Einstein, J. M., & Yeo, G. W. (2020). Direct RNA sequencing enables mA detection in endogenous transcript isoforms at base-specific resolution. RNA , 26(1), 19–28. doi: 10.1261/rna.072785.119
- Maier, K. C., Gressel, S., Cramer, P., & Schwalb, B. (2020). Native molecule sequencing by Nano-ID reveals synthesis and stability of RNA isoforms. Genome Research , 30(9), 1332–1344. doi: 10.1101/gr.257857.119
- Martinez, N. M., & Gilbert, W. V. (2018). Pre-mRNA modifications and their role in nuclear processing. Quantitative Biology , 6(3), 210–227.
- Parker, M. T., Knop, K., Sherwood, A. V., Schurch, N. J., Mackinnon, K., Gould, P. D., … Simpson, G. G. (2020). Nanopore direct RNA sequencing maps the complexity of Arabidopsis mRNA processing and mA modification. eLife , 9, e49658. doi: 10.7554/eLife.49658
- Pratanwanich, P. N., Yao, F., Chen, Y., Koh, C. W. Q., Wan, Y. K., Hendra, C., … Göke, J. (2021). Identification of differential RNA modifications from nanopore direct RNA sequencing with xPore. Nature Biotechnology , 39(11), 1394–1402. doi: 10.1038/s41587-021-00949-w
- Price, A. M., Hayer, K. E., McIntyre, A. B. R., Gokhale, N. S., Abebe, J. S., Fera, A. N. D., … Weitzman, M. D. (2020). Direct RNA sequencing reveals m6A modifications on adenovirus RNA are necessary for efficient splicing. Nature Communications , 11(1), 1–17. doi: 10.1038/s41467-020-19787-6
- Python Language Reference. (2009). Chapman & Hall/CRC Mathematical & Computational Biology. doi: 10.1201/9781584889304.axd
- Shumate, A., Wong, B., Pertea, G., & Pertea, M. (2022). Improved transcriptome assembly using a hybrid of long and short reads with StringTie. PLoS Computational Biology , 18(6), e1009730. doi: 10.1371/journal.pcbi.1009730
- Simpson, J. T., Workman, R. E., Zuzarte, P. C., David, M., Dursi, L. J., & Timp, W. (2017). Detecting DNA cytosine methylation using nanopore sequencing. Nature Methods , 14, 407–410. doi: 10.1038/nmeth.4184
- Smith, A. M., Jain, M., Mulroney, L., Garalde, D. R., & Akeson, M. (2019). Reading canonical and modified nucleobases in 16S ribosomal RNA using nanopore native RNA sequencing. PloS One , 14(5), e0216709. doi: 10.1371/journal.pone.0216709
- Stoddart, D., Heron, A. J., Mikhailova, E., Maglia, G., & Bayley, H. (2009). Single-nucleotide discrimination in immobilized DNA oligonucleotides with a biological nanopore. Proceedings of the National Academy of Sciences of the United States of America , 106(19), 7702–7707. doi: 10.1073/pnas.0901054106
- Stoiber, M., Quick, J., Egan, R., Lee, J. E., Celniker, S., Neely, R. K., … Brown, J. (2017). De novo identification of DNA modifications enabled by genome-guided nanopore signal processing. bioRxiv , doi: 10.1101/094672
- Suzuki, T. (2021). The expanding world of tRNA modifications and their disease relevance. Nature Reviews. Molecular Cell Biology , 22(6), 375–392. doi: 10.1038/s41580-021-00342-0
- Tang, A. D., Soulette, C. M., van Baren, M. J., Hart, K., Hrabeta-Robinson, E., Wu, C. J., & Brooks, A. N. (2020). Full-length transcript characterization of SF3B1 mutation in chronic lymphocytic leukemia reveals downregulation of retained introns. Nature Communications , 11(1), 1438. doi: 10.1038/s41467-020-15171-6
- Taoka, M., Nobe, Y., Yamaki, Y., Sato, K., Ishikawa, H., Izumikawa, K., … Isobe, T. (2018). Landscape of the complete RNA chemical modifications in the human 80S ribosome. Nucleic Acids Research , 46(18), 9289–9298. doi: 10.1093/nar/gky811
- Tavakoli, S., Nabizadehmashhadtoroghi, M., Makhamreh, A., Gamper, H., Rezapour, N. K., Hou, Y. - M., … Rouhanifard, S. H. (2022). Detection of pseudouridine modifications and type I/II hypermodifications in human mRNAs using direct, long-read sequencing. bioRxiv , doi: 10.1101/2021.11.03.467190
- Wang, L., Wang, S., & Li, W. (2012). RSeQC: Quality control of RNA-seq experiments. Bioinformatics , 28(16), 2184–2185. doi: 10.1093/bioinformatics/bts356
- Weilguny, L., De Maio, N., Munro, R., Manser, C., Birney, E., Loose, M., & Goldman, N. (2023). Dynamic, adaptive sampling during nanopore sequencing using Bayesian experimental design. Nature Biotechnology , January. [Epub ahead of print]. doi: 10.1038/s41587-022-01580-z
- Workman, R. E., Tang, A. D., Tang, P. S., Jain, M., Tyson, J. R., Razaghi, R., … Timp, W. (2019). Nanopore native RNA sequencing of a human poly(A) transcriptome. Nature Methods , 16(12), 1297–1305. doi: 10.1038/s41592-019-0617-2
- Zhang, Z., Chen, T., Chen, H. - X., Xie, Y. - Y., Chen, L. - Q., Zhao, Y. - L., … Luo, G. - Z. (2021). Systematic calibration of epitranscriptomic maps using a synthetic modification-free RNA library. Nature Methods , 18(10), 1213–1222. doi: 10.1038/s41592-021-01280-7
Internet Resources
This site has a detailed description of Nanocompore and instructions on how to contribute to the project.

