Using Fluorescent GAP Indicators to Monitor ER Ca2+
Jonathan Rojo-Ruiz, Jonathan Rojo-Ruiz, Cinthia Sánchez-Rabadán, Cinthia Sánchez-Rabadán, Belen Calvo, Belen Calvo, Javier García-Sancho, Javier García-Sancho, Maria Teresa Alonso, Maria Teresa Alonso
Abstract
The endoplasmic reticulum (ER) is the main reservoir of Ca2+ of the cell. Accurate and quantitative measuring of Ca2+ dynamics within the lumen of the ER has been challenging. In the last decade a few genetically encoded Ca2+ indicators have been developed, including a family of fluorescent Ca2+ indicators, dubbed GFP-Aequorin Proteins (GAPs). They are based on the fusion of two jellyfish proteins, the green fluorescent protein (GFP) and the Ca2+-binding protein aequorin. GAP Ca2+ indicators exhibit a combination of several features: they are excitation ratiometric indicators, with reciprocal changes in the fluorescence excited at 405 and 470 nm, which is advantageous for imaging experiments; they exhibit a Hill coefficient of 1, which facilitates the calibration of the fluorescent signal into Ca2+ concentrations; they are insensible to variations in the Mg2+ concentrations or pH variations (in the 6.5-8.5 range); and, due to the lack of mammalian homologues, these proteins have a favorable expression in transgenic animals. A low Ca2+ affinity version of GAP, GAP3 (KD ≅ 489 µM), has been engineered to conform with the estimated [Ca2+] in the ER. GAP3 targeted to the lumen of the ER (erGAP3) can be utilized for imaging intraluminal Ca2+. The ratiometric measurements provide a quantitative method to assess accurate [Ca2+]ER, both dynamically and at rest. In addition, erGAP3 can be combined with synthetic cytosolic Ca2+ indicators to simultaneously monitor ER and cytosolic Ca2+. Here, we provide detailed methods to assess erGAP3 expression and to perform Ca2+ imaging, either restricted to the ER lumen, or simultaneously in the ER and the cytosol. © 2024 The Authors. Current Protocols published by Wiley Periodicals LLC.
Basic Protocol 1 : Detection of erGAP3 in the ER by immunofluorescence
Basic Protocol 2 : Monitoring ER Ca2+
Basic Protocol 3 : Monitoring ER- and cytosolic-Ca2+
Support Protocol : Generation of a stable cell line expressing erGAP3
INTRODUCTION
The endoplasmic (or sarcoplasmic) reticulum (ER/SR) is the main Ca2+ store of the cell. This is the result of three different elements: (1) Ca2+ pumps that accumulate Ca2+ against its concentration gradient; (2) Ca2+ channels that release the stored Ca2+ into the cytosol along its concentration gradient; and (3) luminal Ca2+-binding proteins that buffer the stored Ca2+. Accordingly, the sarco-endoplasmic-reticulum Ca2+ ATPase (SERCA) is located in the ER/SR membrane and, actively pumps Ca2+ from the cytosol into the ER lumen, which can lead to the accumulation of high amounts of Ca2+ against the concentration gradient, up to millimolar levels. On the other hand, Ca2+ release from the ER is evoked by activation of the two main Ca2+ channels: inositol 1,4,5-trisphosphate receptors (IP3Rs) (Berridge, 2016) and ryanodine receptors (RyRs) (Zalk et al., 2007). Finally, the main Ca2+-binding proteins in the ER/SR are calreticulin or calsequestrin (Prins & Michalak, 2011). Proper regulation of intraluminal Ca2+ concentration via these elements is crucial for organelle functions such that an increase or reduction of [Ca2+]ER may trigger processes such as ER stress or apoptosis (Carreras-Sureda et al., 2018).
The ER/SR is a dynamic structure that forms discrete junctions with the plasma membrane and membranes of organelles such as mitochondria or endo-lysosomes (Lam & Galione, 2013). The so-called membrane contact sites are responsible for the exchange of Ca2+ or lipids. The release of Ca2+ from the ER/SR controls many cellular functions, such as muscle contraction, secretion, or synaptic activity (Clapham, 2007). It also controls the Ca2+ entry through the plasma membrane via the so-called store-operated Ca2+ entry (SOCE) mechanism (Putney, 2017). This comprises the Ca2+ channel STIM, present in the ER membrane, and the Orai channel, present in the plasma membrane. Upon cell activation, Ca2+ released from the ER triggers STIM translocation to the plasma membrane, where it interacts with Orai, which opens and activates SOCE. This ubiquitous mechanism is especially relevant for immune function (Vaeth et al., 2020).
Direct monitoring of intraluminal ER Ca2+ is essential for studying the regulation of its dynamics and the associated cellular physiological and pathological processes. Several low Ca2+ affinity chemical probes useful to measure Ca2+ within the ER are available, Furaptra and Fluoraptra being the most utilized (Hofer & Machen, 1993). However, they cannot be targeted to specific cell types or subcellular locations, although some strategies have been successfully developed to trap the probe in the interior of the chosen organelle (Samtleben et al., 2013). In contrast, the genetically encoded Ca2+ indicators (GECIs) can be targeted to any chosen organelle and several have been developed for the ER, including both bioluminescent and fluorescent variants (Alonso et al., 2017). Among the bioluminescent ones, the jellyfish aequorin was the first indicator shown to be functional in the ER (Barrero et al., 1997). The fluorescent indicators are comprised by either one or two fluorescent proteins (e.g., erYC, D1er, erCEPIA, etc.) (Palmer et al., 2004; Suzuki et al., 2014). In recent years, the use of these indicators to monitor ER Ca2+ dynamics has provided invaluable insights to dissect the mechanisms underlying different physiological and pathological conditions.
We have recently developed a new family of GECIs named GAP, for “GFP-Aequorin Protein” (Rodriguez-Garcia et al., 2014). One of the main features of the GAP family is the shift in the fluorescence excitation maximum upon Ca2+ binding, which allows ratiometric measurements. In addition, GAPs are relatively insensible to variations of pH (6.5 to 8.0 pH) and Mg2+ concentrations (range of 0.1-5 mM). Also, they have a Hill coefficient (n) of 1, which simplifies its calibration. The Ca2+ affinity of GAP can be tailored to match with the expected [Ca2+]ER, leading to a low affinity version dubbed GAP3, that has been successfully targeted to the ER of a number of different cell types. Importantly, the fact that GAP only contains jellyfish proteins (GFP and aequorin), makes it a bio-orthogonal indicator, unlikely to interact with endogenous host proteins. This favors functional expression of GAP3 targeted to the ER in several transgenic model organisms (mouse and fly) in vivo with non-apparent off-target effects (Delrio-Lorenzo et al., 2020; Rojo-Ruiz et al., 2021).
Basic Protocol 1 characterizes the subcellular localization of the GAP3 indicator in the ER by immunofluorescence. Basic Protocol 2 describes how to perform imaging of Ca2+ dynamics within the ER upon cellular stimulation with a ubiquitous IP3-coupled agonist. We also provide details to calibrate the erGAP3 fluorescent signal into [Ca2+]ER. Basic Protocol 3 illustrates how to combine ER and cytosolic Ca2+ measurements by simultaneously imaging the signal of GAP3 and a synthetic cytosolic Ca2+ probe. The Support Protocol provides details on how to generate a stable clone expressing GAP3 targeted to the lumen of the ER in an epithelial cell line (ARPE-19).
Basic Protocol 1: DETECTION OF erGAP3 EXPRESSION IN THE ER BY IMMUNOFLUORESCENCE
GAP3 is a chimeric protein consisting of a mutated version of GFP fused to the N-terminus of apoaequorin via a 16-residue linker (Fig. 1A). The GFP variant exhibits 8 substitutions (L15Q, Q80R, F99S, M153T, V163A, I167V, S175G, and D180Y) (Navas-Navarro et al., 2016), with respect to the wild-type GFP, whereas the apoaequorin mutant carries the D24N/D119A double amino acid substitutions (named after the aequorin protein). GAP3 is targeted to the ER by using a targeting strategy widely accepted for this purpose: tagging of calreticulin signal peptide and the ER retention sequence peptide Lys-Asp-Glu-Leu (KDEL), to the N- and C-terminus of GAP3, respectively.
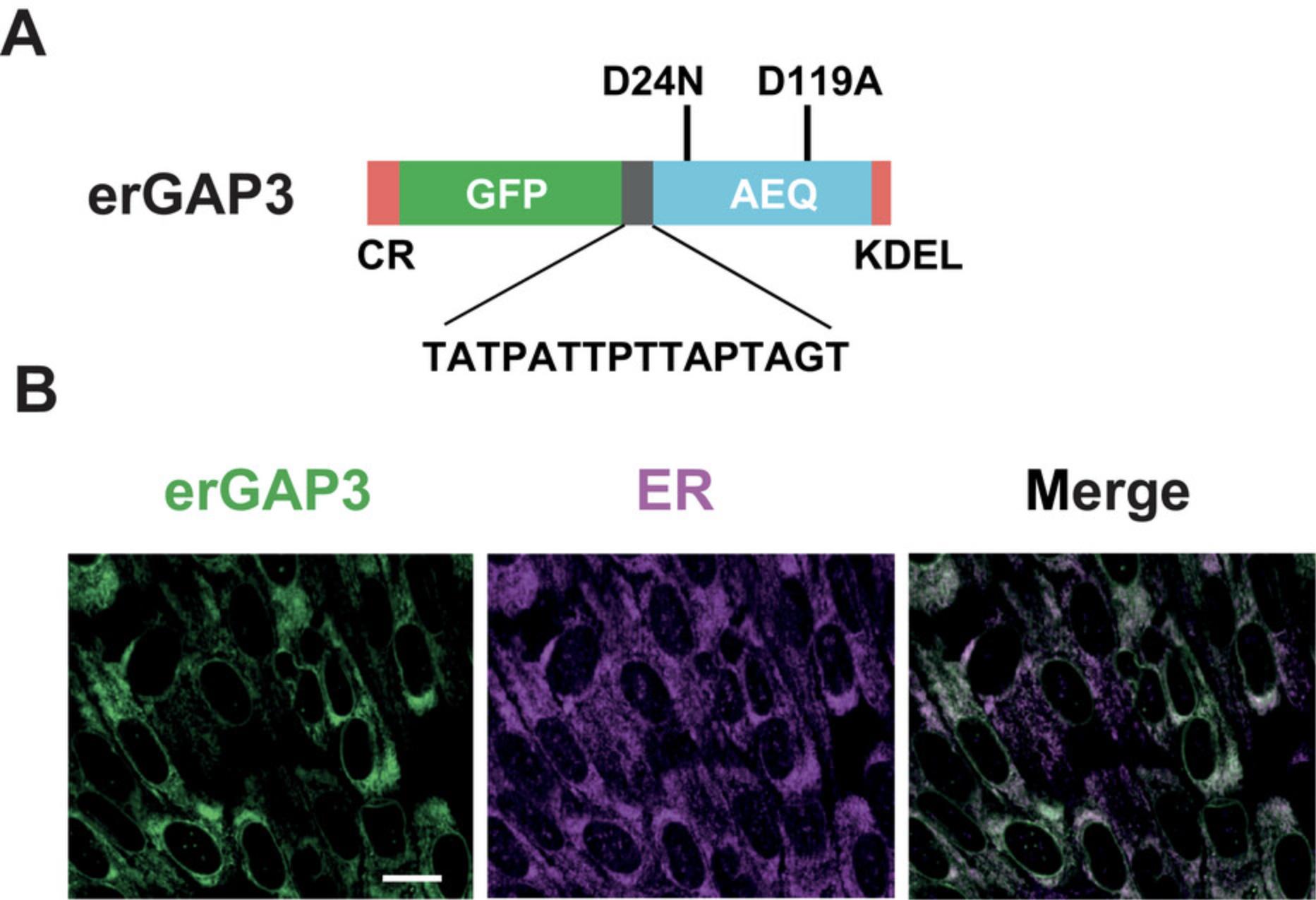
Before performing Ca2+ imaging, it is important to confirm that GAP3 is correctly targeted to the ER. The following protocol discusses the steps to characterize targeting of erGAP3 in cells in which it is stably expressed by comparison between the localization pattern of the indicator and the ER marker calreticulin (Fig. 1B).
Materials
-
ARPE-19 cell line stably expressing erGAP3 (see the Support Protocol)
-
ARPE-19 cell culture medium (see recipe)
-
Phosphate-buffered saline (PBS), pH 7.4 (Gibco, cat. no. 10010-056)
-
4% (v/v) paraformaldehyde (PFA; Sigma-Aldrich, cat. no. 76240)
-
Blocking/permeabilization solution (see recipe)
-
Blocking solution (see recipe)
-
Anti-Calreticulin antibody (Palex Enzo, cat. no. ADI-SPA-600-D)
-
AF568 Goat anti rabbit antibody (Molecular probes, cat. no. A11011)
-
DAPI solution (see recipe)
-
Antifade mounting medium (Vectashield®; Vector Laboratories, cat. no. H-1000)
-
Nail polish
-
Laminar flow hood (Telstar™ Bio II Advance Plus Class II or equivalent)
-
Automatic pipettes (1000, 200, and 20 µl)
-
Poly-L-lysine coverslips (see recipe)
-
4-well cell culture−treated plate (ThermoScientific, cat. no. 176740)
-
Fluorescence tissue culture microscope (Nikon or equivalent)
-
Upright fluorescence microscope (Zeiss Axioplan 2 or equivalent)
-
63×/1.2 W Corr objective (Zeiss)
-
AxioCam MRm camera (12 bits) connected through the software interface Axiovision Rel 4.6.3 (Zeiss)
-
Zeiss ApoTome® module
Immunostaining with an ER marker
1.Seed ∼5 × 104 erGAP3-ARPE-19 cells onto poly-L-lysine-coated 12-mm diameter glass coverslips placed into 4-well plates a day earlier.
2.Remove the culture medium and wash the cells twice, each time with 0.5 ml PBS (5 min/wash).
3.Fix the cells with 0.2 ml/well of 4% PFA fixative and incubate for 15 min at 22°C.
4.Remove PFA and wash the cells three times with 0.5 ml PBS/well during each wash.
5.Aspirate PBS and add 0.5 ml blocking/permeabilization solution. Incubate for 1 hr at 22°C.
6.Aspirate blocking/permeabilization solution. Add 0.2 ml primary antibody solution (rabbit anti-calreticulin) diluted 1:200 (v/v) in blocking solution. Incubate overnight at 4°C.
7.On the following day wash the coverslips three times with blocking solution (0.5 ml/wash), each time for 5 min, to remove unbound primary antibodies.
8.Add 0.2 ml/well secondary antibody solution (AF568 goat anti-rabbit) diluted 1:500 (v/v) in blocking solution. Incubate coverslips in darkness for 1 hr at 22°C.
9.Wash with blocking solution three times (5 min/wash) to remove unbound secondary antibodies.
10.Aspirate blocking solution and add 0.2 ml/well of DAPI solution for nuclear staining. Incubate 5 min at 22°C in darkness.
11.Remove DAPI solution and wash three times, each time with 0.5 ml PBS/well (5 min/wash).
12.Rinse with distilled water to remove salts.
13.Place a small drop (∼2 µl) of VectaShield mounting medium on a glass slide and carefully invert the coverslip and place it on top of the drop without applying pressure.
14.Seal the edges of the coverslip with nail polish to prolong the fluorescence. Leave slides 30 min in darkness on the bench at 20-25°C to allow the nail polish to dry.
15.Turn on the fluorescence microscope. Select 488-nm and 568-nm laser wavelengths of a conventional confocal microscope.
16.Store the slides in the dark up to 6 months at 4°C.
Basic Protocol 2: MONITORING ER Ca2+
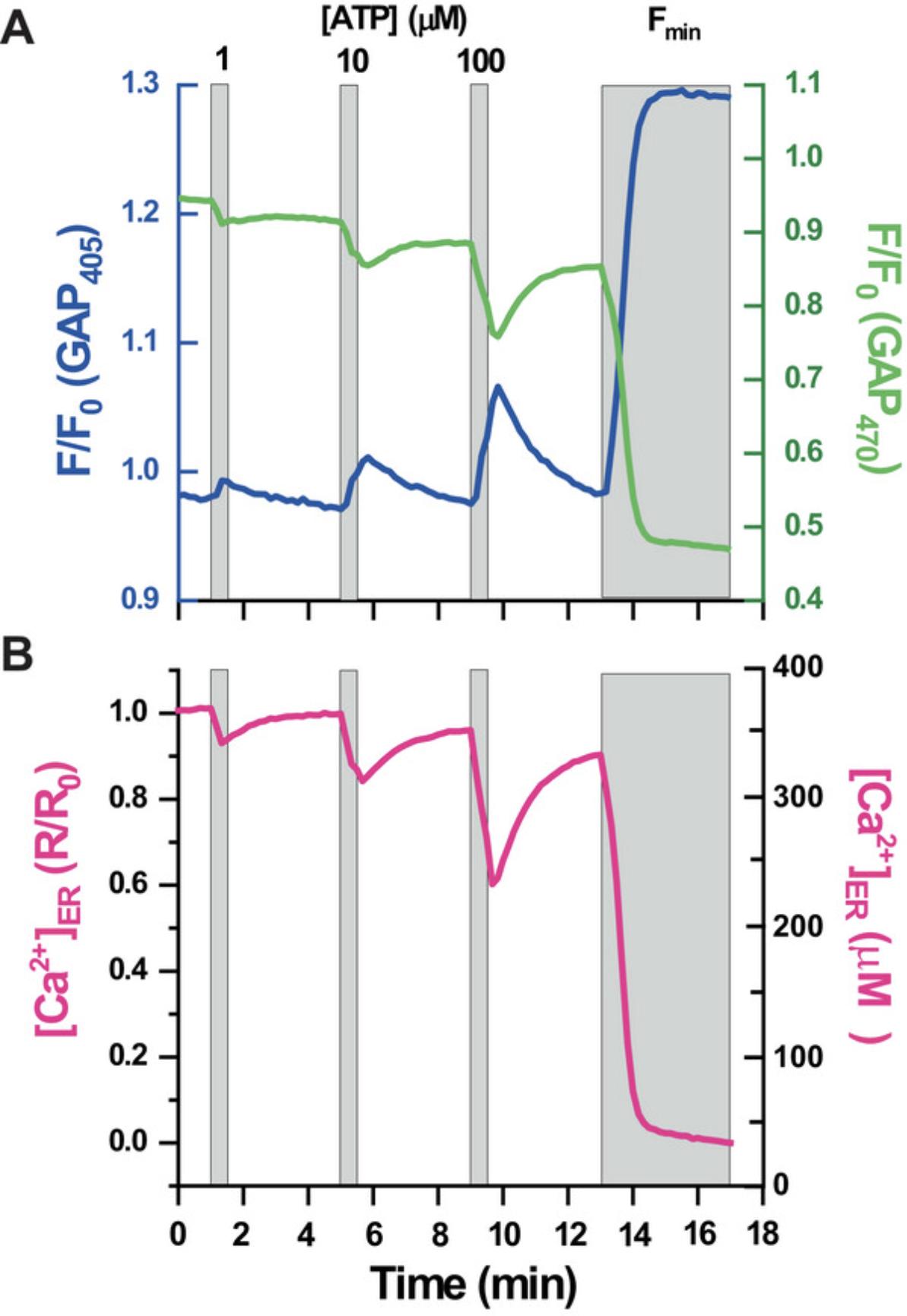
This protocol is intended to explain how to perform live-cell Ca2+ imaging experiments to monitor ER Ca2+ using the indicator GAP3 targeted to the ER. GAP fluorescence shifts its excitation maximum upon Ca2+ binding, allowing ratiometric measurements (Fig. 2). The steps for fluorescence imaging of er-GAP3-expressing ARPE-19 cells using epifluorescence in an upright microscope are discussed. The selected protocol allows monitorization of the fluorescent signals of GAP3 in the lumen of ER both in resting cells and cells stimulated with extracellular ATP. In most cells, ATP is an agonist coupled to the IP3R cascade that provokes a quick drop in the indicator signal as a consequence of the Ca2+ release from the ER resulting in a reduction in the ER Ca2+ content (Fig. 3). To assess for the Ca2+ sensing ability of GAP3, we describe a protocol in which cells are stimulated with three consecutive pulses of increasing concentrations of the agonist. The indicator reports the direct relation of ER Ca2+ release as a function of the concentrations of ATP. At the end of the protocol, a depletion cocktail is added to obtain the minimum value of fluorescence (Fmin) when the indicator is free of Ca2+. This value is required to convert the values of fluorescence into [Ca2+]ER.
![Details are in the caption following the image Spectral properties and sensitivity of erGAP3 to Ca<sup>2+</sup>. (A) Fluorescence excitation spectra of GAP3 (emission set at 520 nm) in the presence of saturating Ca<sup>2+</sup> (10 mM CaCl<sub>2</sub>; magenta trace) and in Ca<sup>2+</sup>-free medium (0.1 mM EGTA; green trace). Measurements were performed with 20 µg recombinant purified protein in a 3-(N-morpholino) propanesulfonic acid (MOPS) solution containing: 140 mM KCl, 1 mM MgCl<sub>2</sub> and 20 mM MOPS-Tris, pH 7.2. (B) In vitro Ca<sup>2+</sup> calibration curve of GAP3. Ca<sup>2+</sup> titrations of GAP3 protein purified from E. coli; curves fit with Hill equation y = (y<sub>max</sub> · [Ca<sup>2+</sup>]<sup>n</sup>) / (K<sub>D</sub><sup>n</sup> + [Ca<sup>2+</sup>]<sup>n</sup>); GAP3 K<sub>D</sub> = 489 µM; Hill coefficient (n) = 1; each point is the mean ± SEM (n = 3). For further details, see (Navas-Navarro et al., 2016). (C) In situ Ca<sup>2+</sup> calibration curve of GAP3. GAP3 K<sub>D</sub> = 539 µM. In situ erGAP3 Ca<sup>2+</sup> calibration was performed in the ER of HeLa cells stably expressing erGAP3. Cells were treated with thapsigargin (1 µM) and permeabilized with digitonin and a cocktail of ionophores to dissipate ionic gradients across the ER membrane. Solutions containing [Ca<sup>2+</sup>] of 0 (EGTA 0.1 mM), 0.1, 0.3, 1, 3, and 10 mM were added. Each point is the mean ± SEM (n = 9 cells). More details in Navas-Navarro (2016).](https://static.yanyin.tech/literature_test/cpz11060-fig-0002-m.jpg)
Additional Materials (also see Basic Protocol 1)
-
EM with 1 mM CaCl2 (EMC; see recipe)
-
EM without CaCl2 (0.5 mM EGTA; EM0Ca; see recipe)
-
ATP working solutions (see recipes)
-
Fmin solution (depletion cocktail; see recipe)
-
75 W xenon arc lamp (Osram, cat. no. XBO75 W/2)
-
Zeiss Axioplan 2 upright microscope equipped with a motorized dichroic turret and an 8-position excitation filter wheel
-
20×numerical aperture (NA) 0.5, long working distance, plastic insulated, water-immersion objective (W-Achroplan; Zeiss cat. no. 440049)
-
Excitation filters installed in the filter wheel: ET402/15× (Chroma, cat. no. 278424) and 470/35DF filter (Omega Optical, cat. no. XF1013)
-
Dichroic mirror 505DRLP (Omega Optical, cat. no. XF2031)
-
Emitter filter 535DF35 (Omega Optical, cat. no. XF3007)
-
Neutral density (ND) filters: ND 1.0 (Omega Optical, cat. no. 142588), ND 0.5 (Omega Optical, cat. no. 164526) and ND 0.3 (Omega Optical, cat. no. 2620425)
-
AxioCam MRm camera (Zeiss; 12 bits Ser. N°. 1 30 030879 r20 0445-554)
-
Axiovision Rel 4.6.3 software (Zeiss)
-
8-valve perfusion system (Automate Scientific; ValveLink8.2 perfusion Controller)
1.Seed ∼5 × 104 cells onto poly-L-lysine-coated 12-mm diameter glass coverslips in individual wells of a 4-well plate the day before performing the experiment.
2.Pour the different solutions to be perfused in bottles or tubes: 500 ml Extracellular Medium with CaCl2 (EMC) in a bottle; 50 ml of each of the solutions with the three [ATP] (1, 10, and 100 µM), and 100 ml of Fmin solution. Connect the valves and purge the perfusion system.
3.Check that the perfusion system operates properly.
4.Place the glass coverslip with the seeded ARPE-19 cells into the perfusion chamber. Connect the perfusion chamber for imaging.
5.Perfuse cells with EMC.
6.Focus on the plane where the cells are localized by using the brightfield illumination.
7.Choose a field of view by using the fluorescence illumination.
8.Select the sampling rate of image acquisition and exposure time for 405 and 470 nm illumination using the appropriate software.
9.Start time-lapse data acquisition.
10.To record resting ER Ca2+, perform data acquisition at least for 1 min with EMC perfusion before perfusing any cell stimuli.
11.At min 1, switch to 1 μM ATP solution and perfuse for 30 s.
12.Return to EMC solution and perfuse up to min 5.
13.At min 5 and at min 9, repeat steps 11 and 12 with 10 and 100 μM solutions of ATP.
14.At min 13, perfuse “Fmin solution” to calibrate the signal of erGAP3.Record during 4-6 min until the fluorescent signal is stable.
15.At min 17, stop data acquisition and save data as TIFF file.
Data Analysis of erGAP3 fluorescence imaging
16.Open ImageJ software.
17.Open the files.
18.Adjust brightness and contrast to better visualize the image, if necessary (select Image ˃ Adjust ˃ Brightness/Contrast).
19.Scroll through the stack file to have a general view of the cell responses along the experiment. Repeat this for each channel.
20.Duplicate image stack and select regions of interest (ROIs) in the chosen cells of the duplicated image.
21.Subtract image background for each channel (select Process ˃ Subtract Background).
22.Limit the grayscale range values to a range containing relevant information, by selecting a lower threshold value which excludes background and noise (select Image > Adjust > threshold).
23.Repeat step 22 for each channel.
24.Calculate the ratio between the fluorescence signal at 470 (F470) and 405 nm (F405) [select Process > Image Calculator > Operation: Divide Ch (F470) by Ch (F405)]. You can also use Image Calculator Plus plugin downloaded from https://imagej.net/ij/plugins/index.html: Plugins > Calculator Plus > Operation: Divide. Save the file as TIFF images for further analysis.
25.Calculate the average fluorescence intensity values for each ROI in F470, F405 and ratio time stacks [select one of the stacks, e.g., ratio stack > ROI Manager (with the previously selected ROIs), check the dialog box “Show All” > more > Multi Measure: check the dialog boxes “Measure all slices” and “one row per slice” > OK. Perform the same steps for the other two stacks].
26.Copy the results _ mean gray value data_ of the ROIs and open it with a software for graphing and analysis, e.g., excel.
27.Plot graphics for ratio, F470 and F405 values.
28.Average the ratio of data obtained in all cells.
29.Calculate R0 by averaging the first 5-10 R data. Divide ratio data set by R0 to obtain R/R0.
Basic Protocol 3: MONITORING ER AND CYTOSOLIC Ca2+
Simultaneous monitoring of cytosolic and ER Ca2+ dynamics obviously provides a more complete picture of the cellular Ca2+ dynamics than monitoring exclusively Ca2+ in one of the compartments. erGAP3 can be used in combination with a cytosolic Ca2+ probe like fura-2 (Tsien, 1999) (KD for Ca2+, 135 nM at 20°C or 224 nM at 37°C) or rhod-3 (KD for Ca2+, 570 nM at 20°C). These indicators can be easily loaded into the cells by incubating them with its acetoxymethyl ester (AM; Fig. 4A) derivatives. The AM groups mask the four (or more) carboxylates of the probes, permitting its diffusion through the plasma membrane and, once in the cytosol, are susceptible to cytoplasmic esterases that hydrolyze the ester groups, regenerating the acidic form that is trapped inside the cells. Fura-2, one of the most popular Ca2+ indicators, has an excitation peak at 360 nm, which shifts to 340 nm and increases in amplitude upon binding of Ca2+ (Fig. 4B). Both, Ca2+-free and Ca2+-bound fura-2 show an emission peak at 510 nm. This allows ratiometric measurements of cytosolic Ca2+, such that fluorescence increases at 340 nm, and decreases at 380 nm when Ca2+ binds to the indicator. Rhod-3 is a proprietary red indicator with improved properties with respect to its predecessor rhod-2, showing improved fluorescence emission and no internalization into organelles. It displays an excitation peak at 540 nm and a maximal emission at 580 nm (Fig. 4C). We present here a well-established protocol to monitor SOCE, consisting in inhibiting the SERCA pump by incubating the cells with a specific SERCA inhibitor in absence of extracellular Ca2+. This passively empties the ER Ca2+ store monitored by erGAP3 indicator, and provokes the activation the SOCE, monitored by an increase of cytosolic Ca2+ reported by cytosolic probes, when adding back Ca2+ to the extracellular media (Putney, 2017). The results are obtained in a HeLa cell line stably expressing erGAP3 loaded with a cytosolic Ca2+ indicator (fura-2 or rhod-3) (Fig. 5). The choice of using fura-2 instead of rhod-3 allows to obtain more accurate ratiometric data for cytosolic Ca2+, but compromises the ER Ca2+ measurements, as only one GAP wavelength (470 nm) is imaged. On the contrary, the combination of using rhod-2 with erGAP3 allows to use single wavelength cytosolic and ratiometric ER Ca2+ measurements. If the aim of the experiment is obtaining accurate resting ER Ca2+ or calibrated [Ca2+]ER, then, erGAP3 must be expressed as ratio and combined with rhod-3.Alternatively, if accurate cytosolic measurements are a priority, then the fura-2/erGAP3 should be preferred.
![Details are in the caption following the image Structure and properties of cytosolic Ca<sup>2+</sup> indicators. (A) Structure of fura-2 AM. (B) Excitation spectrum of fura-2 and right shift of the peak upon binding of Ca<sup>2+</sup>. Solutions of EGTA-K<sub>2</sub>-H<sub>2</sub> and EGTA-Ca-K<sub>2</sub> were blended to yield the final [Ca<sup>2+</sup>] indicated in nM. Measurements were performed in saline solutions containing 130 mM KCl, 20 mM NaCl, 1.1 mM MgCl<sub>2</sub>, and 10 mM MOPS-K, pH 7.05. (Image from Alonso, 1990; Grynkiewicz et al., 1985). (C) Excitation and emission spectra of rhod-3. [Image reproduced with permission from Thermo Fisher Scientific (https://www.thermofisher.com/order/catalog/product/R10145)].](https://static.yanyin.tech/literature_test/cpz11060-fig-0004-m.jpg)
![Details are in the caption following the image SOCE activation monitored as coordinated cytosolic and ER Ca<sup>2+</sup> responses. (A) HeLa cells stably expressing erGAP3 were loaded with fura-2 AM. Cells were treated for 10 min with TBH (10 µM) in EM0Ca medium prior to starting the experiment. SOCE is triggered by addition of CaCl<sub>2</sub> (1 mM). [Ca<sup>2+</sup>]<sub>C</sub> is imaged by fura-2 at two wavelengths (340 and 380 nm) and recorded as R/R<sub>0</sub>, where R is the ratio of F<sub>340</sub>/F<sub>380</sub> and [Ca<sup>2+</sup>]<sub>ER</sub> (erGAP3) is expressed as normalized F<sub>470</sub> (F/F<sub>0</sub>). Each trace corresponds to the average obtained from 75 cells from the same optical field. This experiment is representative for three repetitions performed at different days. (B) HeLa cells stably expressing erGAP3 were loaded with rhod-3 AM. Cells were treated with thapsigargin (200 nM) in EM0Ca medium prior to imaging. [Ca<sup>2+</sup>]<sub>C</sub> is imaged by rhod-3 at 550 nm and normalized as F/F<sub>0</sub>, and [Ca<sup>2+</sup>]<sub>ER</sub> (erGAP3) is represented as R/R<sub>0</sub>, where R is the ratio (F<sub>470</sub>/F<sub>405</sub>). Each trace corresponds to the average obtained from 115 cells from the same optical field and this figure is representative for at least four independent experiments.](https://static.yanyin.tech/literature_test/cpz11060-fig-0005-m.jpg)
Additional Materials (also see Basic Protocol 2)
-
Fura-2 AM (Thermo Fisher Scientific, cat. no. F1201)
-
Rhod-3 AM (Thermo Fisher Scientific, cat. no. R1045)
-
2,5-di-tert-butylhydroquinone (TBH) in EM0Ca solution (see recipe)
-
TBH in EMC solution (see recipe)
-
Thapsigargin in EM0Ca solution (see recipe)
-
Excitation filters installed in the filter wheel for Fura-2 imaging: 340AF15 (XF1093, Omega optic) and 380/AF15 filter (XF1093, Omega optic)
-
Cube set filter 15 for rhod-3 (Zeiss, ca. no. 488015-0000-000): 546/12 excitation filter, FT580 dichroic mirror, and LP590 emitter filter
Monitoring fura-2 and erGAP3
1.Follow steps 1-3 from Basic Protocol 2.
2.Add 0.2 μl of 2 mM fura-2 stock solution into 0.2 ml EMC medium to obtain a fresh 2 µM Fura-2 AM working solution. Place this solution in a well of a 4-well plate.
3.Insert a 12-mm glass coverslip on which the stably expressing erGAP3 cells (in this protocol, HeLa cells) have been seeded to the well containing fura-2 AM.
4.Incubate for 45 min at 22°C at slow speed in a shaker.
5.Replace medium with 0.5 ml of TBH in EM0Ca solution.
6.Incubate for 10 min at 22°C.
7.Transfer the coverslip to the perfusion cell chamber on the microscope stage and start the perfusion with TBH in EM0Ca solution.
8.Focus on the plane where the cells are localized using the brightfield illumination and choose a field using the fluorescence illumination.
9.Select the correct channels to record fura-2 (340 and 380 nm) and erGAP3 (470 nm). Make sure that cells are loaded with fura-2.
10.Select the sampling rate for image acquisition and exposure time for 340, 380, and 470 nm illumination using the appropriate software.
11.Adjust the minimal excitation exposure times for both indicators to obtain an acceptable SNR.
12.Start data acquisition.
13.At min 2, switch to TBH in EMC solution.
14.Stop acquisition at min 12.
15.Save data as a TIFF file.
Monitoring rhod-3 and erGAP3
16.Follow steps 1-3 from Basic Protocol 2.
17.Add 0.4 µl of 1 mM rhod-3 stock solution into 0.2 ml EMC medium to obtain a fresh 2 µM rhod-3 AM working solution. Place this solution in a well of a 4-well plate.
18.Insert a 12-mm glass coverslip onto which the stably expressing erGAP3-HeLa cells have been seeded to a well containing 2 µM rhod-3.
19.Incubate for 45 min at 22°C with slow speed in a shaker.
20.Replace medium with 0.5 ml of 200 nM thapsigargin solution in EM0Ca.
21.Incubate for 10 min.
22.Remove medium and add 0.5 ml EM0Ca to wash out thapsigargin.
23.Transfer the coverslip to a perfusion cell chamber on the microscope stage and start the perfusion with EM0Ca solution.
24.Focus on the plane where cells are localized using the brightfield illumination and choose a field using the fluorescence illumination.
25.Select the correct channels to record rhod-3 (546 nm) and erGAP3 (405 and 470 nm). Make sure that cells are loaded with rhod-3.
26.Select the sampling rate of image acquisition and exposure time for 405, 470, and 546 nm illumination.
27.Adjust the minimal excitation exposure times for both indicators to obtain an acceptable SNR.
28.Start data acquisition.
29.At min 2, switch to EMC solution.
30.Stop acquisition at min 12.
31.Save data as a TIFF file.
Data Analysis of cytosolic and erGAP3 fluorescence imaging
Perform the same routine analysis as detailed in steps 16-29 of Basic Protocol 2 for the analysis of the ER Ca2+signal but taking into account that you are now working at three different wavelengths.
Fura-2 and erGAP3 recordings
Analyze the ratio for fura-2 and the fluorescence at 470 nm for erGAP3 using ImageJ software. Obtain the ratio between the fura-2 fluorescence signal at 340 (F340) and 380 nm (F380) and plot R/R0 (Fig. 5A, magenta trace). For erGAP3 signal, analyze the individual wavelength, at 470 nm and represent F/F0 (Fig. 5A, green trace).
Rhod-3 and erGAP3 recordings
Analyze the ratio for erGAP3 and the fluorescence at 546 nm for rhod-3 using ImageJ software. Obtain the ratio between the erGAP3 fluorescence signal at 470 (F470) and 405 nm (F405) and plot R/R0 (Fig. 5B, green trace). For rhod-3 signal, analyze the individual 546 nm wavelength and represent F/F0 (Fig. 5B, magenta trace).
Support Protocol: GENERATION OF A STABLE CELL LINE EXPRESSING erGAP3
There are multiple reasons to generate a cell line that stably express erGAP3: (1) it avoids constantly having to transfect the corresponding cDNA, thus, saving time and resources; (2) it permits cell selection that expresses the transgene with specific properties, like a good SNR, and no signs of precipitates; (3) it facilitates the reproducibility of the results among different experiments since all cells used are identical; and (4) it permits studies of cellular communication (e.g., intercellular wave propagation), for which it is essential that all cells express the indicator.
In this article we present protocols using two stable cell lines, one derived from ARPE-19 (Basic Protocols 1 and 2) and another from HeLa cells (Basic Protocol 3). HeLa cells are one of the most widely used cell lines and ARPE-19 cells are widely used to model human retinal epithelium functions, drug testing or intercellular Ca2+ wave spreading. Both cell lines were first transiently transfected and then we selected cells in which erGAP3 was stably integrated into the genome, such that all the rest of cells that did not take the transgene up or that maintained it episomally, were eliminated by adding a selectable marker. Under selection, these cells continuously express the transgene even following cell division. Here we describe a protocol to generate a stable monoclonal ARPE-19 cell line expressing erGAP3.The protocol to generate a HeLa cell line expressing erGAP3 is practically identical.
Additional Materials (also see Basic Protocols 1-3)
-
pcDNA3.erGAP3 (Addgene, cat. no. 78118)
-
ARPE-19 cells (ATCC, cat. no. CRL-2302)
-
Lipofectamine 2000 (Thermo Fisher Scientific, cat. no. 11668027)
-
G418 solution (see recipe)
-
0.05% Trypsin-EDTA (Gibco, cat. no. 25300-096)
-
100-mm Petri dishes (Corning 100-mm × 20-mm Style Dish, cat. no. 430167)
-
Sorter BD FAcSaria
-
24-well plates (VWR International, cat. no. 30623-116)
-
96-well plates (Thermo Fisher Scientific, cat. no. 136101)
NOTE : In order to choose the optimal concentration of G418, prepare a killing curve for G418.Use, for instance concentrations between 100, 250, 500, 750, and 1000 μg/ml. Choose the minimal concentration that produces death of all cells after 7 days of treatment. In the experiments described below, we used 800 μg/ml. For maintenance of stable clones, concentration of G418 can be lowered to 200 μg/ml.
Transient transfection and antibiotic selection
1.Pre-warm ARPE-19 complete cell culture medium in a culture bath at 37°C.
2.Change medium of the cell culture (grown up to 60% confluence) right before transfection.
3.On day 0, transfect cells with 3 μg erGAP3 plasmid in a 100-mm plate by addition of lipofectamine 2000 following the manufacturer's guidelines. Incubate cells for 2 days without adding G418.
4.On day 2, add 800 μg/ml of G418.Monitor cell death, morphology and GFP fluorescence under the fluorescence microscope during the following 3 days.
5.On day 6, trypsinize the cells, collect them, and proceed to their sorting by excitation with a 488-nm laser. Collect the live GFP-positive cells.
6.Seed the cells again in a 100-mm petri dish with culture medium containing G418.
7.After 7-10 days of antibiotic selection, all the untransfected or transiently transfected cells will be dead and individual GFP-positive colonies should be visible.
8.Inspect the plate for colony formation and mark and select the colonies to be isolated according to their GFP fluorescent intensity and characteristic ER localization pattern.
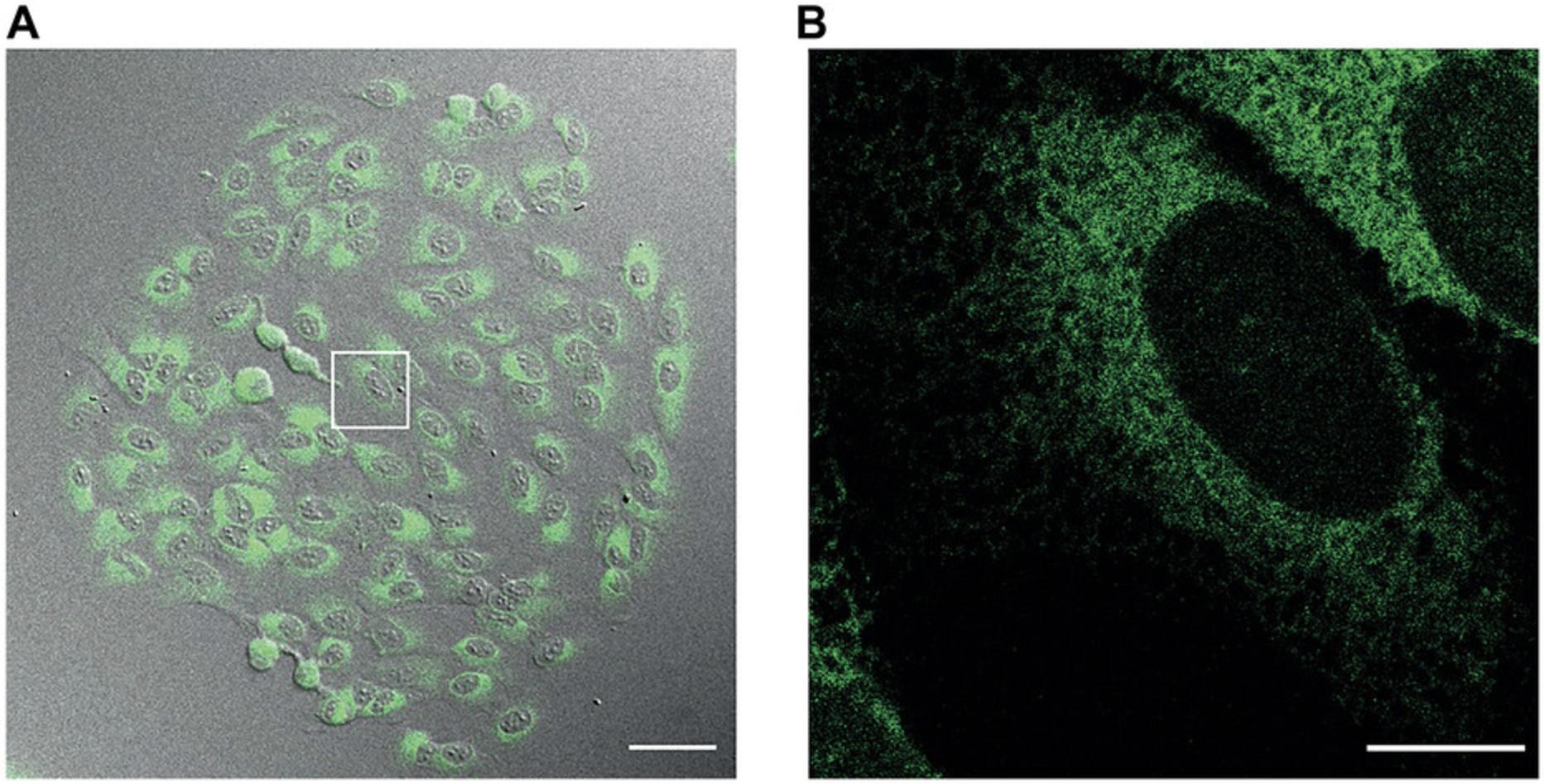
9.Isolate colonies from the plates by trypsinizing cells under the fluorescence microscope by adding 10 µl trypsin to each colony.
10.Transfer each colony to individual wells in a 24-well plate containing selective medium.
11.Expand the colonies to provide enough cells for freezing.
12.Choose one colony to generate monoclonal cell lines.
Generation of monoclonal cells
13.Add 50 μl culture medium in each well of a 96-well plate and incubate for 20 min at 37°C.
14.Trypsinize the selected colony from step 11.
15.Perform limiting dilution by counting cells and resuspending them with complete growth medium at a density of 1.6 cells/ml (Freshney, 2010) (https://knowledge.lonza.com/downloadasset.ashx?assetId=28243). Then, seed 50 μl of this solution in each well of a 96-well plate. This will ensure a single cell in some wells.
16.Inspect and count number of cells per well 18 to 24 hr after plating. Select wells that have only one cell and discard the wells that have more than one cell or none at all.
17.Monitor GFP fluorescence by using a fluorescence inverted microscope and check cell morphology. Ensure that the pattern of fluorescence is characteristic for the ER (Fig. 6).
18.Inspect selected cells by using brightfield illumination to ensure that the cell morphology appears healthy, well attached to the plate, with no intracellular vacuoles or bubbles. Alternate to fluorescence illumination to confirm that the localization of erGAP3 has a characteristic ER pattern without any precipitates (Fig. 6). If all cells are fluorescent, grow them to confluency.
19.If some cells are not fluorescent, go back to step 14 and resuspend at a density of 8 cells/ml. Repeat steps 15 to 18.
20.Trypsinize the cells and expand them first to a 24-well plate, and then to a 100-mm petri dish.
21.Freeze the cells first at −80°C for a maximum of 7 days and then in liquid nitrogen for several years.
REAGENTS AND SOLUTIONS
Use Milli-QR distilled water in all recipes in which it is included.
ARPE-19 cell culture medium
- Fetal bovine serum (FBS)
- Penicillin and streptomycin
To a bottle of 500 ml Dulbecco's Modified Eagle Medium F-12 Nutrient Mixture (DMEM/F-12 (1:1) (1×) + GlutaMAXTM supplement; Gibco, cat. no. 31331-028) add 55 ml heat-inactivated fetal bovine serum (FBS; Cytiva, cat. no. SV30160.03), and 5.5 ml penicillin/streptomycin (Sigma-Aldrich, cat. no. P4458). Store up to 6 months at 4°C.
ATP working solutions
Make 25 ml of 100 µM ATP by taking 25 µl of the ATP-Mg stock solution (see recipe) and dilute into 25 ml EMC medium. From this solution make two serial dilutions of 10 and 1 µM of ATP. Prepare fresh immediately before use.
ATP-Mg stock solution, 0.1 M
Dissolve 5.51 g Adenosine 5′-triphosphate disodium salt hydrate (ATP; Sigma-Aldrich, cat. no. A3377) in 10 ml of a 1 M MgCl2 solution (see recipe). Adjust pH to 7.0 with NaOH. Bring to 0.1 L final volume with distilled water. Divide into 10-ml aliquots. Store frozen up to 24 months at −20°C.
Blocking solution
To prepare 50 ml, take 45 ml PBS and add 5 ml goat serum [ThermoFisher, cat. no. 16210064; final concentration 10% (v/v)]. Prepare fresh immediately before use.
Blocking/permeabilization solution
Add 0.25 ml Triton X-100 [TX-100; Merck, cat. no. 108603; final concentration 0,5% (v/v)] to 50 ml blocking solution. Prepare fresh immediately before use.
CaCl2 solution, 1 M
Dissolve 73.51 g CaCl2 dihydrate (Merck, cat. no. 1.02382.0500) in 0.5 L distilled water to make a 1 M stock solution. Divide into 10-ml aliquots. Store frozen up to 24 months at −20°C.
DAPI, stock and working solutions
To obtain a 1 mg/ml DAPI (Invitrogen, cat. no. D1306) stock solution, dissolve the content of one vial (10 mg) in 10 ml distilled water. Divide into 1-ml aliquots. Store frozen and protected from light up to 6 months at −20°C. To prepare 5 ml DAPI working solution, add 5 µl DAPI stock solution to 5 ml PBS (final concentration 1 µg/ml). Prepare fresh immediately before use.
D-Glucose solution, 1 M
Dissolve 198.17 g D-glucose (D-Glucose monohydrate, Merk, cat. no. 108342) in 1 L distilled water to make a 1 M stock solution. Divide into 50-ml aliquots. Store frozen up to 12 months at −20°C.
EGTA solution, 1 M
Dissolve 190.75 g EGTA [Ethylene Glycol-Bis (β-aminoethyl ether)-N ,N ,N ',N '-tetraacetic acid; Sigma-Aldrich, cat. no. E4378] in 0.5 L distilled water to make a 1 M stock solution. Adjust to pH 8 with NaOH for complete solubilization. Divide into 50-ml aliquots. Store frozen up to 12 months at −20°C.
EM with 1 mM CaCl2 (EMC)
- To 1 L EM add 1 ml of 1 M CaCl2. Store up to 6 months at 4°C.
- EM without CaCl2 (EM0Ca)
To 1 L EM add 0.5 ml of 1 M EGTA (0.5 mM final concentration). The final volume of EMC and EM0Ca depends on the specific protocol. Calculate volumes beforehand taking into account the perfusion speed and the different solutions of the protocol. For the experiment described in Basic Protocol 1 we typically prepare 1 L of EMC and 500 ml of EM0Ca solutions. Store up to 6 months at 4°C.
Extracellular medium (EM), 10×
- To prepare 1 L add the following :
- 84.74 g of NaCl (Merck, cat. no. 106404; 1.45 M final concentration)
- 3.73 g of KCl (Merck, cat. no. 104936; 50 mM final concentration)
- 23.83 g of Na-HEPES [4-(2-Hydroxyethyl) piperazine-1-ethanesulfonic Acid; Sigma-Aldrich, cat. no. H3375; 100 mM final concentration]
- 2.03 g of MgCl2 × 6H2O (Sigma-Aldrich, cat. no. M9272; 10 mM final concentration)
- Dissolve in 0.99 L distilled water
- Adjust pH to 7.6 with NaOH
- Bring to a final volume of 1 L with distilled water
- Store at 4°C or frozen for 12 months
Extracellular medium (EM), 1×
Take 0.1 L 10× EM solution and 10 ml of a 1 M D-glucose solution (see recipe); 10 mM final concentration. Bring to a final volume of 1 L with distilled water. The pH of this solution should be 7.4 and it is not necessary to confirm it. This solution can be stored up to 1 month at 4°C.
Fmin solution (depletion cocktail)
To prepare 100 ml, take 99.8 ml EM0Ca solution (see recipe for EMC solution) and add 0.1 ml of a 0.1 M ATP stock solution (0.1 mM final concentration) and 0.1 ml TBH stock solution (10 µM final concentration). Prepare fresh immediately before use.
Fura-2 stock solution, 2 mM
Dissolve the content of a tube (50 µg) into 24.9 µl DMSO (Corning, cat. no. 25-950-CQC; 2 mM final concentration). Store frozen up to 6 months at −20°C.
NOTE : DMSO should be of the best quality. Maintain fura-2 stock protected from light.
G418 solution
Prepare 100 mg/ml G418 (Gibco, cat. no. 11811-03) stock in H2O. Solutions should be filter sterilized prior to storage up to 24 months at 2°C to 8°C. Solutions can also be stored at −20°C.
MgCl2 solution, 1 M
Dissolve 10.10 g MgCl2 dihydrate in 0.05 L distilled water to make a 1 M stock solution. Divide into 10-ml aliquots. Store up to 24 months frozen at −20°C.
Poly-L-lysine coverslips
Prior to use, coat sterile coverslips with sterile poly-L-lysine (0.1% (w/v), prepared in distilled water) for 1 hr at 22°C. Rinse the coverslips with sterile distilled water three times, 5 min each wash and then, allow them to air dry for 1-2 hr in a sterile environment. Store up to 12 months at 4°C.
Rhod-3 stock solution, 1 mM
Dissolve the content of a tube (50 µg) in 44.5 µl DMSO to obtain a 1 mM stock solution. Store up to 6 months frozen at −20°C.
NOTE : Maintain rhod-3 stock protected from light.
TBH in EM0Ca solution
Add 50 µl 10 mM TBH stock solution (see recipe) to 50 ml EM0Ca solution (see recipe for EM0Ca solution). Prepare fresh immediately before use.
TBH in EMC solution
Add 50 µl 10 mM TBH stock solution (see recipe) to 50 ml EMC solution (see recipe). Prepare fresh immediately before use.
TBH stock solution, 10 mM
Dissolve 44.46 mg 2,5-di-tert-butylhydroquinone (TBH; Aldrich, cat. no. 11,297-6) in 20 ml DMSO (10 mM final concentration). Divide into 0.5-ml aliquots. Store frozen at −20°C up to 1 year.
Thapsigargin in EM0Ca solution
To prepare 1 ml, take 1 ml EM0Ca solution (see recipe for EMC solution) and add 0.2 µl of a 1 mM thapsigargin stock (see recipe; 200 nM final concentration). Prepare fresh immediately before use. Keep it on ice and protected from light during the working day.
Thapsigargin stock solution, 1 mM
Dissolve 1 mg thapsigargin (Calbiochem, cat.no. 586005) in 1.536 ml DMSO (1 mM final concentration). Divide into 0.1-ml aliquots. Store frozen at −20°C up to 1 year. Protect from light.
COMMENTARY
Background Information
The choice of a Ca2+ indicator with the optimal KD is essential for accurate measurements in the selected organellar environment. The first functional measurements reported by a GECI targeted to the ER were provided by the bioluminescent aequorin (Barrero et al., 1997). The first fluorescent ER GECI, the yellow chameleons (erYCs), were based on Förster Resonance Energy Transfer (FRET) and consisted of two fluorescent proteins and calmodulin as a Ca2+-binding protein (Miyawaki et al., 1997). Since then, a flurry of iterative evolution has led to the development of optimized ER Ca2+ indicators enabling specific monitoring of intraluminal Ca2+ signals at high spatial and temporal resolution with improved sensitivity and brightness (Alonso et al., 2017; Hill et al., 2014; Suzuki et al., 2016).
During the last decade, our laboratory has developed a family of Ca2+ indicators dubbed GAPs, which includes some low affinity members that conform to the high [Ca2+] expected in the ER. The prototype variant, dubbed GAP1, displayed a KD of 17 μM and preserved all the main properties of the original high affinity GAP (mentioned in the article Introduction) (Rodriguez-Garcia et al., 2014). In the ER, where the estimated [Ca2+] values are 200-1000 μM, GAP1 was able to detect large changes of luminal Ca2+ induced by strong mobilizers, but it was less sensitive to detect small fluctuations. Further optimization led to GAP2, that contained the mutations D119A and D24N in the aequorin moiety and exhibited a KD for Ca2+ of 407 μM (Navas-Navarro et al., 2016). Finally, we developed another variant, dubbed GAP3, by optimizing the codon usage for mammalian expression and re-engineering several residues (L15Q, I167V, S175G, and D180Y) in the GFP moiety, to increase its fluorescence intensity. The KD of GAP3 is 489 μM and its dynamic range is 3-4, so that GAP3 is the optimal choice for targeting to the ER (erGAP3) and measuring Ca2+ in its lumen.
The erGAP3 indicator combines several desirable features in a low-affinity fluorescent GECI. First, it is ratiometric, with two excitation peaks (at 405 and 470 nm), at which fluorescence intensities change inversely upon Ca2+ binding (Fig. 2A). The ratiometric indicators are advantageous over single wavelength indicators, because ratioing corrects spurious fluorescent changes due to photobleaching, focal plane shift, differences in expression levels, tissue thickness or other conditions that may result in fluorescence fluctuations unrelated to Ca2+ changes. Ratiometric detection minimizes the detrimental effect of movements and allows reliable imaging in preparations displaying a considerable movement like migrating or motile cells which exhibit shape changes over time. Also, in long term imaging experiments, especially in vivo , ratiometric detection is preferred. Finally, to accurately record steady-state [Ca2+]ER ratiometric sensors are required and, at rest, they are usually brighter than single fluorescent ones. erGAP3 is bright and easily detected in resting cells (unstimulated) at both wavelengths.
Second, the dynamic range of erGAP3 in the ER lumen is ∼3-4. The dynamic ranges and Ca2+ affinities may vary considerably depending on the environment. Often, Ca2+ indicators are calibrated in vitro but, when used in situ , their dynamic range is usually reduced. For that reason, it is advisable to calibrate the sensor in the target organelle. The KD and dynamic range of erGAP3 are practically identical in vitro and in situ , in the ER lumen (Fig. 2B and C).
Third, erGAP3 possesses a Hill coefficient (n) of 1.0 (possibly due to the presence of only one functional EF hand) while preserving the fluorescence signaling mechanism. It exhibits a linear relationship between changes in Ca2+ concentration and fluorescence within the calcium range of interest. The combination of being ratiometric together with the Hill coefficient of 1.0 enables to yield quantitative data of [Ca2+]ER.
Fourth, GAP is practically insensitive to variations of pH (in the 6.5 to 8.5 pH range) or in [Mg2+] (in the 0.1 to 5 mM range). Although pH sensitivity is not such a serious problem in the ER lumen, it could be relevant in acidic organelles with high Ca2+ content, such as the Golgi apparatus, for instance.
Fifth, the GAP family of Ca2+ indicators have no endogenous partners since it is derived from the fusion of GFP and aequorin, two jellyfish proteins with no mammalian homologues. This fact minimizes the possibility of GAP interacting with endogenous partners and contrasts with other sensors based on endogenous proteins such as calmodulin. Besides this, the Ca2+ binding unit, aequorin, was the first Ca2+ probe to be established, and since the 60's it has been extensively used, even at high concentrations, with no reports of disturbances due to binding to endogenous mammalian partners (Blinks et al., 1978; Shimomura et al., 1962). This orthogonal nature of the GAP family explains why the erGAP3 indicator is functional in most cells tested, including an array of primary cells such as astrocytes, neurons, pituitary cells, pancreatic islet cells or dendritic cells. A further proof for its biocompatibility is its correct performance upon expression in the context of transgenic organisms. We have generated a number of different lines expressing erGAP3 in transgenic organisms, both in flies and mice, in which we have addressed the role of ER Ca2+ for specific functions such as hormone secretion in pituitary cells (Rojo-Ruiz et al., 2021) or muscle contraction in skeletal muscle (Delrio-Lorenzo et al., 2020).
Most of the cytosolic Ca2+ signals originate in the ER. However, in many cases cytosolic Ca2+ signals only roughly correlate with ER Ca2+ signals. Therefore, direct luminal ER Ca2+ measurements are required to quantitatively assess the ER Ca2+ dynamics. Basic Protocol 2 allows to measure Ca2+ dynamics specifically in the ER by imaging the fluorescence changes of erGAP3 in response to a general agonist like ATP, due to Ca2+ release from the ER via IP3Rs. The amplitude of the ER Ca2+ responses are proportional to the [ATP] used, and this is an indication of the sensibility of the Ca2+ indicator.
Ratiometric erGAP3 measurements allow to estimate resting luminal [Ca2+]ER in various pathophysiological conditions (Mekahli et al., 2011). One example for such a pathological condition are mutations in genes associated with Alzheimer's disease where the ER Ca2+ passive leak is increased, and concomitantly the [Ca2+]ER is reduced (Palmer et al., 2004). In turn, a decrease in the [Ca2+]ER may produce ER stress and/or the unfolding protein response (UPR) (Carreras-Sureda et al., 2018).
Another field for application of these techniques is the excitation-contraction in skeletal or cardiac muscle. In this case, ratiometric detection is recommended to correct for the movement associated to the contraction. Also, the selected indicator should exhibit fast kinetics for Ca2+ binding, sufficient for reliably monitoring the fast contraction. Several ER Ca2+ indicators have been employed for studying muscle contraction, including some detecting ratiometric emission (Jiménez-Moreno et al., 2010; Rudolf et al., 2006; Sztretye et al., 2011). Calcium-induced calcium release (CICR) is an important mechanism during which the entry of Ca2+ provokes an increase in the cytosolic Ca2+ and a release of Ca2+ from the ER, which, in turn, activates IP3Rs or RyRs. Therefore, CICR is a representative example where simultaneous Ca2+ monitoring in the two compartments can give reliable insight into this process (Rodríguez-Prados et al., 2020).
Basic Protocol 2 can also be used to study Ca2+ oscillations. These can be evoked by release of ER Ca2+ using a submaximal concentration of IP3-producing agonists or by Ca2+ entry from the extracellular medium through the plasma membrane (Schulte et al., 2022). These processes can be better studied by simultaneous monitoring of cytosolic and ER Ca2+ dynamics (Basic Protocol 3) since the cytosolic Ca2+ oscillations are coordinated with those in the ER (Rodríguez-Prados et al., 2020). One ubiquitous mechanism of Ca2+ entry triggered by the emptying of ER Ca2+ and aimed to replenish the ER with Ca2+, SOCE, is monitored here using Basic Protocol 3. In addition to cytosolic Ca2+, erGAP3 may be used in combination with other indicators which are spectrally compatible and may be targeted to other organelles like mitochondria or the nucleus (e.g., using a red indicator for mitochondria).
Finally, another application of [Ca2+]ER monitoring is high-throughput screening (HTS) for drug discovery. By using a stably expressing erGAP3 cell line (Support Protocol), different screening approaches can be developed as tools for specific drug discovery or for identification of compounds that interfere with ER Ca2+ functions (Murayama et al., 2018).
Critical Parameters
There are a number of critical parameters that one should pay attention to:
- The expression of erGAP3 should be high enough to guarantee a good SNR. If the GFP signal is too dim this will give a high background. The DNA of the erGAP3 must be of highest quality since this is crucial for transfection efficiency.
- The choice of the optimal excitation and emission filters is essential to obtain the maximum yield of the GAP3. In principle, an inverted or an upright fluorescent microscope are both suitable for imaging. Also, 40× or 60× objectives can be used to obtain a higher spatial resolution. The use of objectives with a high numerical aperture is highly recommended.
- In an imaging system, an optimized perfusion system is essential to guarantee a fast exchange of agonist solutions (to stimulate cells and to wash out the stimuli) which allows to address the reversibility of the responses. In addition to the valves, a critical parameter of the perfusion system is the volume of the cell chamber that should be as small as possible because the speed of the solution exchange may influence the temporal rise and decay of the fluorescence signals. The valves can be controlled remotely or manually. In studies focused on the steady state [Ca2+]ER, the perfusion system is not essential as the Fmin can be obtained by manually adding the depletion cocktail.
- In theory, all fluorescent indicators suffer from photobleaching when they are continuously excited. To minimize this effect, it is important to excite cells with the lowest intensity of light possible, and this is especially relevant for lengthy protocols. Also, when the protocol includes applications that last several minutes (e.g., perfusion of the depletion cocktail) the shutter may be closed. Confocal microscopy may also be used with this protocol, although in our hands the responses are smaller. When applied the depletion cocktail to obtain Fmin the amplitude of the signal is lower compared to that observed in a non-confocal microscope. This might be due to the high intensity of the excitation laser, which quenches part of the signal.
- It is crucial to confirm that the observed decrease in ER Ca2+ results from the corresponding changes in the individual fluorescence of erGAP3, namely, a decrease in fluorescence at 470 nm and an increase at 405 nm. If only one channel changes, or both channels change in the same direction (decrease or increase), this fluctuation may be attributable to other spurious reasons, unrelated to Ca2+.
- In order to convert the observed ratios for erGAP3 into [Ca2+]ER it is essential to obtain the Rmin at the end of each experiment. To determine Rmin, we add a depletion cocktail that guarantees a complete emptying of the Ca2+ from the ER. The cocktail consists of a general IP3 producing agonist like ATP together with the SERCA inhibitor TBH in the absence of Ca2+. The time required to reach Rmin might vary for each cell type, so to reach an accurate Rmin it is important to wait till the fluorescent signal does not change further.
- As previously mentioned, Ca2+ indicators can act as Ca2+ buffers and therefore, may distort the physiological changes they intend to report (McMahon & Jackson, 2018). Thus, it is important to recognize the impact of Ca2+ indicators on the results, especially when imaging cytosolic Ca2+. The practical consequences of the Ca2+ buffering effect is double: it dampens the amplitude of the Ca2+ transient and, it blurs the spatial signal. So, it is advisable to minimize the sensor's buffering capacity by using the lowest possible concentration of the Ca2+ probe (in Basic Protocol 3, fura-2 or rhod-3) to provide a sufficient SNR. In practice, a compromise must be reached between small interference in the physiological calcium signaling and optimization of the SNR.
- The experiments are mostly performed at temperatures of 22°-25°C, but in some preparations (e.g., pancreatic Langerhans islets or cardiac cells) cells should be maintained at 37°C. In those cases, all the solutions must be pre-heated in a water bath and heated with an Inline Solution Heater.
Troubleshooting
Table 1 shows the common problems with the protocols, their causes, and potential solutions.
| Problem | Possible cause | Solution |
|---|---|---|
| Poor fluorescence signal of erGAP3 at rest. | No or low expression of erGAP3. |
Check the plasmid DNA in an agarose gel for any signs of degradation. Increase the amount of DNA used for transfection. Change the plasmid to use a stronger promoter. Check that the excitation filters are optimal. Check the healthiness of the cells. |
| Poor fluorescence signal of erGAP3 at rest. | No specific signal due to high background. | Increase acquisition time and/or increase gain. Shorten the protocol. |
| None, small or slow fluorescence changes upon addition of stimulus. |
The volume of the perfusion chamber is too large. The perfusion flow speed is too slow, or it has stopped. |
Change the perfusion cell chamber. Increase perfusion flow speed. Check that perfusion is not blocked. |
| Continuous drift of fluorescent signal at both wavelengths. | Low erGAP3 expression and/or high light exposure (gain either too high or too long). | Increase erGAP3 expression and/or reduce gain of exposure. |
| Spotty erGAP3 expression. |
Possible aggregation of erGAP3. No homogeneous distribution within the ER. |
Reduce the amount of DNA used to transfect cells. Select a low-expressor clone or use a weaker promoter. |
| Image suddenly disappears. |
Possibly perfusion problems due to bubbles. Cells detach from the coverslip because culture is too dense. |
Discard last frames. Check solutions and perfusion system. Seed cells at lower density. |
| Sudden punctuate appearance of erGAP3 fluorescence upon addition of certain solutions. | Cells start to die. | The stimulus solution is toxic for the cell. Minimize concentration/time of stimulus or use an alternative. |
| No fura-2 (or rhod-3) signal (Basic Protocol 3). | Fura-2 (or rhod-3) is not loaded into the cells. |
Check the probe stock and/or make a new working solution. Check that cell culture, especially primary cells, is healthy with no vacuoles or other signs of aging. |
| None or low rhod-3 fluorescence signal (Basic Protocol 3). | Decay of rhod-3. | Rhod-3 is very unstable in the working solution. Make a new working solution. |
Statistical Analysis: (optional)
One of the advantages of performing Ca2+ imaging experiments is to analyze the cell-to-cell heterogeneity in the Ca2+ responses. Thus, it is important to analyze the performance of each selected cell by first plotting the individual traces of ER (and cytosolic, if that is the case) Ca2+ and then, inspect the cell-to-cell variations that may occur. Then, calculate the mean of the cells with the same phenotype, first in the same microscope field and then, of several independent days. Finally, perform statistics within the various phenotypic groups found.
Understanding Results
GAP3 is a fluorescent Ca2+ indicator consisting of GFP and aequorin, and unlike bioluminescent aequorin, it does not require reconstitution with any cofactor. It is a low Ca2+ affinity derivative of its GAP predecessor generated by substituting two amino acid residues (D24N and D119A) in the EF-hand sites I and III of the aequorin protein (Kendall, Sala-Newby, et al., 1992). In order to target GAP3 to the lumen of the ER, a standard strategy was applied that has been validated for most of the Ca2+ indicators developed to date (reviewed by (Hill et al., 2014; Pendin et al., 2017)). It consists in appending a consensus ER targeting signal (calreticulin signal peptide) to the N-terminal end of GAP3, and a specific sequence that ensures retrieval from post-ER compartments by retrograde transport mechanisms to the C-terminus (Kendall, Dormer, et al., 1992; Fig. 1A). The most broadly used ER-targeting retrieval sequence is the lysine-aspartate-glutamate-leucine (KDEL) tetrapeptide. A reported variant of this strategy to target a Ca2+ sensor to the SR utilizes the calsequestrin signal peptide (Sztretye et al., 2011). A completely different strategy, mostly used for bioluminescent aequorin-based biosensors, consists of the fusion with the N-terminal domain of a protein (e.g., an immunoglobulin) that specifically binds to BIP protein, an ER resident protein (Montero et al., 1995).
The specific targeting of GAP3 to the ER should be confirmed in the selected cellular model by unambiguous localization of the fluorescent indicator using high resolution imaging and co-localization with known ER markers by immunofluorescence (Basic Protocol 1). Figure 1B shows the results of such an experiment performed in the stable cell line ARPE-19, a suitable model to image Ca2+ due to the large size and flat appearance of cells, and their interesting Ca2+ signaling mechanisms (Leybaert & Sanderson, 2012). The green fluorescence of GAP3 displays a bright reticulate staining, throughout the cytosol reaching from the perinuclear area to the periphery of the cell and is excluded from the nucleus. Despite the fact that this ARPE-19 cell line is a clonal cell line, the erGAP3 expression levels are quite variable among cells. Importantly, GAP3 co-localizes with the endogenous ER marker calreticulin in all cells. Moreover, the expression is relatively homogeneous throughout the cytosol of all cells and there are no signs of aggregation or punctuate precipitates, which might reflect non-functional molecules of the Ca2+ indicator.
In the protocols presented here, both cell lines (ARPE-19 and HeLa) are stably transfected, which, in general, facilitates to perform the experiments. But, in principle, any cellular model may be used in which erGAP3 expression is obtained through transient transfection, viral transduction or by transgenesis. The erGAP3 plasmid is available in Addgene or upon request.
Like its predecessor GAP, the excitation spectrum of GAP3, shown in Figure 2A, maintains two peaks, at 405 and 470 nm, and is sensitive to Ca2+. Thus, binding to Ca2+ (magenta trace) provokes a red shift in the spectrum, reducing the fluorescence at 405 and increasing it at 470 nm. The emission spectrum exhibits a unique peak at 510 nm which is not modified by the addition of Ca2+. In vitro calibration of GAP3 with Ca2+ shows a KD of 489 µM, very appropriate for measuring luminal [Ca2+]ER (Fig. 2B). One common problem encountered with Ca2+ sensors affect the KD, which can considerably vary among different environments or compartments. Moreover, the dynamic range (DR) value obtained with recombinant purified protein in vitro often shrinks in the targeted organelle, in situ. Thus, it is highly recommended to calibrate the sensor in the final destination, in this case, the ER lumen. In general, Ca2+ calibration requires the dissipation of transmembrane potentials and active transport mechanisms across the membrane to avoid the formation of concentration gradients and this can be particularly difficult to achieve. We have established a protocol for efficient calcium equilibration of ER (Navas-Navarro et al., 2016). As shown in Figure 2C the results obtained in the ER for the DR and KD in situ are very similar to those obtained in vitro. The DR value of GAP3 is around 3 for both cases.
To test functionality of erGAP3 we followed Basic Protocol 2 during which we stimulated cells for 30 s with ATP, a general agonist coupled to IP3 production, which provoked an activation of IP3R and, thus, Ca2+ release from the ER. This can be visualized as a fast decline in the fluorescent signal of the indicator (Fig. 3). The addition of ATP provoked reciprocal changes at the two wavelengths, in such a way that fluorescence excited at 405 nm increased and decreased at 470 nm (Fig. 3A). The use of ratio F470/F405 (R) amplifies the changes recorded in individual channels (Fig. 3B). In addition, it allows correcting the fluorescence drift (especially at 470 nm) observed along the 18 min that the protocol shown lasts. The drop in R is observed during the presence of the stimulus. We applied three consecutive pulses of ATP, using increasing concentrations, from 1 up to 100 µM. The R dropped upon each ATP pulse, reflecting a reduction in the [Ca2+]ER, and recovered its basal levels as soon as the stimulus was washed out. This shows that the Ca2+ signals are reversible, indicating that the Ca2+ that is released from the ER into the cytosol is pumped back into the ER by the SERCA. The ER Ca2+ releases are proportional to [ATP], demonstrating that erGAP3 is sensitive enough to detect submaximal Ca2+ reductions in the ER lumen. At the end of the experiment, a solution of Fmin, also named depletion cocktail , is applied to provoke a complete discharge of the ER Ca2+ stores. This includes addition of a high [ATP] (100 µM) in Ca2+-free medium along with the reversible SERCA inhibitor TBH. ATP is very convenient for this purpose because it is a ubiquitous IP3-coupled agonist, but other agonists may also be used. In ARPE-19 the high [ATP] provokes a 40% reduction (R/R0 falls from 1.0 to 0.6) in the ER Ca2+ content although this value can vary among cell types. The SERCA inhibitor causes a reduction of the ER Ca2+ signal due to a passive exit of Ca2+ from the ER and the resulting slow depletion of the ER calcium. The combination of ATP and TBH in absence of external Ca2+ guarantee that Rmin is reached. It is convenient to represent the normalized ratios (R/R0).
The conversion of R into [Ca2+]ER is performed according to the following equation:
Using equation 1, resting [Ca2+]ER can be calculated from the ratios obtained in cells at rest, before addition of the stimuli.
Basic Protocol 3 describes the simultaneous monitoring of cytosolic and ER Ca2+. We combined erGAP3 with any of the two cytosolic indicators, either fura-2 or rhod-3. Both are loaded into the cells as acetoxymethyl ester (AM) derivatives, as they are membrane-permeable (Fig. 4A). Ca2+-free fura-2 exhibits an excitation peak at 360 nm, which shifts to 340 nm upon binding of Ca2+ (Fig. 4B). Both Ca2+-bound and the Ca2+-free fura-2 have their emission peak at 510 nm. This allows ratiometric measurements of cytosolic Ca2+, such that fluorescence increases at 340 nm, and it decreases at 380 nm when Ca2+ binds to the indicator. In contrast, rhod-3 is a red intensiometric indicator, with only one excitation peak at 540 and emission at 580 nm (Fig. 4C).
In this protocol we have addressed the activation of SOCE for two main reasons. First, this mechanism is a ubiquitous process for Ca2+ entry through the plasma membrane, secondary to the Ca2+ emptying of the ER. Second, there is a well-established protocol to visualize its activation as a transient increase in the cytosolic Ca2+. It consists in depleting the ER Ca2+ stores by incubating the cells for 10 min with a SERCA inhibitor (either the reversible TBH or the irreversible thapsigargin) in the absence of extracellular Ca2+ and then, applying 1 mM Ca2+ to provoke the sudden entry of Ca2+ through the Orai channels located in the plasma membrane.
In the first protocol, cytosolic Ca2+ is imaged with fura-2 at two wavelengths (340 and 380 nm) and ER Ca2+ is imaged using erGAP3 recording only at 470 nm (Fig. 5A). Thus, cytosolic Ca2+ is represented as R/R0 and ER Ca2+ as F/F0. Cells are incubated with TBH for 10 min prior to imaging. At the beginning of the recording, in the absence of extracellular Ca2+ the fluorescent signal of fura-2 is low, indicating that cytosolic [Ca2+] is low and the re-addition of Ca2+ (1 mM) results in a fast and transient increase of cytosolic Ca2+, as a consequence of the rapid Ca2+ entry through the Orai channels into the cytosol. This peak slowly (∼5 min) returns to the basal level despite the continuous presence of extracellular Ca2+, indicating that the cytosolic Ca2+ is being removed by clearance mechanisms, located at the plasma membrane or at other endomembranes. The Ca2+ dynamics in the ER are very similar to those of the cytosol. The ER Ca2+ store is empty at the beginning of the experiment and the sudden entry of Ca2+ into the cytosol provokes a parallel and relatively modest and transient increase of Ca2+ into the ER (less than 20% change in the F/F0) which quickly returns to the original level. This indicates a passive entry of Ca2+ into the ER, probably due to the presence of a leak channel operating in reverse mode followed by the exit of Ca2+, probably through the same channel.
In the second protocol we used rhod-3 instead of fura-2 (Fig. 5B). In this case, rhod-3 is imaged at only one wavelength (560 nm) and represented as F/F0, whereas erGAP3 is imaged in both channels (405 and 470 nm) and represented as R/R0. In this case cells are treated with the irreversible SERCA blocker thapsigargin for 10 min before the beginning of the experiment. Results are very similar to those described in Figure 5A.
Time Considerations
Basic protocols 1, 2, and 3 each take typically 3 days provided that a stably expressing cell line is used. If transient transfection is required, another 2 days must be added. The Support Protocol can take 1-2 months.
Acknowledgments
This study was supported by grants from the Ministerio de Economía y Competitividad (BFU2017-83066-P and PID2020-116086RB-I00), from the Consejería de Educación de la Junta de Castilla y León (GR175) and from the Programa Estratégico “Instituto de Biología y Genética Molecular (IBGM)”, Escalera de Excelencia, Junta de Castilla y León (CLU-2019-02). We thank Rebeca Llorente for assistance with confocal microscope; Jesús Fernández, and Iris López and former members Miriam García and Carla Rodríguez for expert technical help. We also thank Dr. Schimmang for helpful comments on the manuscript.
Author Contributions
Jonathan Rojo-Ruiz : Conceptualization; data curation; formal analysis; investigation; methodology; software; visualization; writing—review and editing. Cinthia Sanchez-Rabadan : Data curation; formal analysis; investigation; visualization; writing—review and editing. Belen Calvo : Data curation; formal analysis; investigation; visualization; writing—review and editing. Javier García-Sancho : Conceptualization; funding acquisition; investigation; supervision; writing—review and editing. Maria Teresa Alonso : Conceptualization; funding acquisition; project administration; supervision; writing—original draft; writing—review and editing.
Conflict of Interest
The authors declare no conflict of interest.
Open Research
Data Availability Statement
All the row data that support the figures presented in this article are available from the corresponding author upon reasonable request.
Literature Cited
- Alonso, M. T. (1990). Entrada y liberación de Calcio en plaquetas estimuladas. Evidencia de un canal operado por receptor [Unpublished doctoral Dissertation]. University of Valladolid.
- Alonso, M. T., Rojo-Ruiz, J., Navas-Navarro, P., Rodriguez-Prados, M., & Garcia-Sancho, J. (2017). Measuring Ca2+ inside intracellular organelles with luminescent and fluorescent aequorin-based sensors. Biochimica et Biophysica Acta , 1864(6), 894–899. https://doi.org/10.1016/j.bbamcr.2016.12.003
- Arnold, L. W., & Lannigan, J. (2010). Practical issues in high-speed cell sorting. Current Protocols in Cytometry , 51, 1.24.1–1.24.30. https://doi.org/10.1002/0471142956.cy0124s51
- Barrero, M. J., Montero, M., & Alvarez, J. (1997). Dynamics of Ca2+ in the endoplasmic reticulum and cytoplasm of intact HeLa cells. A comparative study. Journal of Biological Chemistry , 272(44), 27694–27699. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=9346910
- Berridge, M. J. (2016). The inositol trisphosphate/calcium signaling pathway in health and disease. Physiological Reviews , 96(4), 1261–1296. https://doi.org/10.1152/physrev.00006.2016
- Blinks, J. R., Mattingly, P. H., Jewell, B. R., van Leeuwen, M., Harrer, G. C., & Allen, D. G. (1978). [31]Practical aspects of the use of aequorin as a calcium indicator: Assay, preparation, microinjection, and interpretation of signals. Methods in Enzymology , 57, 292–328. https://doi.org/10.1016/0076-6879(78)57033-X
- Carreras-Sureda, A., Pihán, P., & Hetz, C. (2018). Calcium signaling at the endoplasmic reticulum: Fine-tuning stress responses. Cell Calcium , 70, 24–31. http://www.ncbi.nlm.nih.gov/pubmed/29054537
- Clapham, D. E. (2007). Calcium signaling. Cell , 131(6), 1047–1058. http://www.ncbi.nlm.nih.gov/pubmed/18083096
- Delrio-Lorenzo, A., Rojo-Ruiz, J., Alonso, M. T., & García-Sancho, J. (2020). Sarcoplasmic reticulum Ca2+ decreases with age and correlates with the decline in muscle function in Drosophila. Journal of Cell Science , 133(6), jcs240879. https://doi.org/10.1242/jcs.240879
- Freshney, R. I. (2010). Cloning and selection. In R. I. Freshney (Ed.), Culture of Animal Cells (pp. 207–225). https://doi.org/10.1002/9780470649367.ch13
- Grynkiewicz, G., Poenie, M., & Tsien, R. Y. (1985). A new generation of Ca2+ indicators with greatly improved fluorescence properties. Journal of Biological Chemistry , 260(6), 3440–3450. https://www.sciencedirect.com/science/article/pii/S0021925819836414?via%3Dihub
- Hill, J. M., De Stefani, D., Jones, A. W., Ruiz, A., Rizzuto, R., & Szabadkai, G. (2014). Measuring baseline Ca2+ levels in subcellular compartments using genetically engineered fluorescent indicators. Methods in Enzymology , 543, 47–72. https://doi.org/10.1016/b978-0-12-801329-8.00003-9
- Hofer, A. M., & Machen, T. E. (1993). Technique for in situ measurement of calcium in intracellular inositol 1,4,5-trisphosphate-sensitive stores using the fluorescent indicator mag-fura-2. Proceedings of the National Academy of Sciences USA , 90(7), 2598–2602. https://doi.org/10.1073/pnas.90.7.2598
- Jiménez-Moreno, R., Wang, Z.-M., Messi, M. L., & Delbono, O. (2010). Sarcoplasmic reticulum Ca2+ depletion in adult skeletal muscle fibres measured with the biosensor D1ER. Pflugers Archiv: European Journal of Physiology , 459(5), 725–735. http://www.ncbi.nlm.nih.gov/pubmed/20069312
- Kendall, J. M., Dormer, R. L., & Campbell, A. K. (1992). Targeting aequorin to the endoplasmic reticulum of living cells. Biochemical and Biophysical Research Communications , 189(2), 1008–1016. https://doi.org/10.1016/0006-291x(92)92304-g
- Kendall, J. M., Sala-Newby, G., Ghalaut, V., Dormer, R. L., & Campbell, A. K. (1992). Engineering the CA2+-activated photoprotein aequorin with reduced affinity for calcium. Biochemical and Biophysical Research Communications , 187(2), 1091–1097. https://doi.org/10.1016/0006-291X(92)91309-E
- Lam, A. K., & Galione, A. (2013). The endoplasmic reticulum and junctional membrane communication during calcium signaling. Biochimica Et Biophysica Acta , 1833(11), 2542–2559. https://doi.org/10.1016/j.bbamcr.2013.06.004
- Leybaert, L., & Sanderson, M. J. (2012). Intercellular Ca2+ waves: Mechanisms and function. Physiological Reviews , 92(3), 1359–1392. https://doi.org/10.1152/physrev.00029.2011
- McMahon, S. M., & Jackson, M. B. (2018). An inconvenient truth: Calcium sensors are calcium buffers. Trends in Neurosciences , 41(12), 880–884. https://doi.org/10.1016/j.tins.2018.09.005
- Mekahli, D., Bultynck, G., Parys, J. B., De Smedt, H., & Missiaen, L. (2011). Endoplasmic-reticulum calcium depletion and disease. Cold Spring Harbor Perspectives in Biology , 3(6). https://doi.org/10.1101/cshperspect.a004317
- Miyawaki, A., Llopis, J., Heim, R., McCaffery, J. M., Adams, J. A., Ikura, M., & Tsien, R. Y. (1997). Fluorescent indicators for Ca2+ based on green fluorescent proteins and calmodulin. Nature , 388(6645), 882–887. https://doi.org/10.1038/42264
- Montero, M., Brini, M., Marsault, R., Alvarez, J., Sitia, R., Pozzan, T., & Rizzuto, R. (1995). Monitoring dynamic changes in free Ca2+ concentration in the endoplasmic reticulum of intact cells. EMBO Journal , 14(22), 5467–5475. https://doi.org/10.1002/j.1460-2075.1995.tb00233.x
- Murayama, T., Kurebayashi, N., Ishigami-Yuasa, M., Mori, S., Suzuki, Y., Akima, R., Ogawa, H., Suzuki, J., Kanemaru, K., Oyamada, H., Kiuchi, Y., Iino, M., Kagechika, H., & Sakurai, T. (2018). Efficient high-throughput screening by endoplasmic reticulum Ca2+ measurement to identify inhibitors of ryanodine receptor Ca2+-release channels. Molecular Pharmacology , 94(1), 722–730. https://doi.org/10.1124/mol.117.111468
- Navas-Navarro, P. (2016). Diseño de un nuevo sensor de Ca2+ y su aplicación a los organulos intracelulares ex vivo e in vivo University of Valladolid]. https://uvadoc.uva.es/handle/10324/18635
- Navas-Navarro, P., Rojo-Ruiz, J., Rodriguez-Prados, M., Ganfornina, M. D., Looger, L. L., Alonso, M. T., & Garcia-Sancho, J. (2016). GFP-aequorin protein sensor for ex vivo and in vivo imaging of Ca2+ dynamics in high-Ca2+ organelles. Cell Chemical Biology , 23(6), 738–745. https://doi.org/10.1016/j.chembiol.2016.05.010
- Palmer, A. E., Jin, C., Reed, J. C., & Tsien, R. Y. (2004). Bcl-2-mediated alterations in endoplasmic reticulum Ca2+ analyzed with an improved genetically encoded fluorescent sensor. Proceedings of the National Academy of Sciences USA , 101(50), 17404–17409. https://doi.org/10.1073/pnas.0408030101
- Pendin, D., Greotti, E., Lefkimmiatis, K., & Pozzan, T. (2017). Exploring cells with targeted biosensors. Journal of General Physiology , 149(1), 1–36. https://doi.org/10.1085/jgp.201611654
- Prins, D., & Michalak, M. (2011). Organellar calcium buffers. Cold Spring Harbor Perspectives in Biology , 3(3), a004069. https://doi.org/10.1101/cshperspect.a004069
- Putney, J. W. (2017). Store-operated calcium entry: An historical overview. Advances in Experimental Medicine and Biology , 981, 205–214. https://doi.org/10.1007/978-3-319-55858-5_9
- Rodriguez-Garcia, A., Rojo-Ruiz, J., Navas-Navarro, P., Aulestia, F. J., Gallego-Sandin, S., Garcia-Sancho, J., & Alonso, M. T. (2014). GAP, an aequorin-based fluorescent indicator for imaging Ca2+ in organelles. Proceedings of the National Academy of Sciences USA , 111(7), 2584–2589. https://doi.org/10.1073/pnas.1316539111
- Rodríguez-Prados, M., Rojo-Ruiz, J., García-Sancho, J., & Alonso, M. T. (2020). Direct monitoring of ER Ca2+ dynamics reveals that Ca2+ entry induces ER-Ca2+ release in astrocytes. Pflugers Archiv: European Journal of Physiology , 472(4), 439–448. https://doi.org/10.1007/s00424-020-02364-7
- Rojo-Ruiz, J., Navas-Navarro, P., Nuñez, L., García-Sancho, J., & Alonso, M. T. (2021). Imaging of endoplasmic reticulum Ca2+ in the intact pituitary gland of transgenic mice expressing a low affinity Ca2+ indicator. Frontiers in Endocrinology , 11(1117). https://doi.org/10.3389/fendo.2020.615777
- Rudolf, R., Magalhaes, P. J., & Pozzan, T. (2006). Direct in vivo monitoring of sarcoplasmic reticulum Ca2+ and cytosolic cAMP dynamics in mouse skeletal muscle. Journal of Cell Biology , 173(2), 187–193. https://doi.org/10.1083/jcb.200601160
- Samtleben, S., Jaepel, J., Fecher, C., Andreska, T., Rehberg, M., & Blum, R. (2013). Direct imaging of ER calcium with targeted-esterase induced dye loading (TED). Journal of Visualized Experiments , 75, e50317. https://doi.org/10.3791/50317
- Schulte, A., Bieniussa, L., Gupta, R., Samtleben, S., Bischler, T., Doering, K., Sodmann, P., Rittner, H., & Blum, R. (2022). Homeostatic calcium fluxes, ER calcium release, SOCE, and calcium oscillations in cultured astrocytes are interlinked by a small calcium toolkit. Cell Calcium , 101, 102515. https://doi.org/10.1016/j.ceca.2021.102515
- Shimomura, O., Johnson, F. H., & Saiga, Y. (1962). Extraction, purification and properties of aequorin, a bioluminescent protein from the luminous hydromedusan, Aequorea. Journal of Cellular Physiology , 59, 223–239. https://doi.org/10.1002/jcp.1030590302
- Suzuki, J., Kanemaru, K., & Iino, M. (2016). Genetically encoded fluorescent indicators for organellar calcium imaging. Biophysical Journal , 111(6), 1119–1131. https://doi.org/10.1016/j.bpj.2016.04.054
- Suzuki, J., Kanemaru, K., Ishii, K., Ohkura, M., Okubo, Y., & Iino, M. (2014). Imaging intraorganellar Ca2+ at subcellular resolution using CEPIA. Nature Communications , 5, 4153. https://doi.org/10.1038/ncomms5153
- Sztretye, M., Yi, J., Figueroa, L., Zhou, J., Royer, L., Allen, P., Brum, G., & Ríos, E. (2011). Measurement of RyR permeability reveals a role of calsequestrin in termination of SR Ca2+ release in skeletal muscle. The Journal of General Physiology , 138(2), 231–247. http://www.ncbi.nlm.nih.gov/pubmed/21788611
- Tsien, R. Y. (1999). Monitoring cell calcium. In E. Carafoli & C. B. Klee (Eds.), Calcium as a Cellular Regulator (pp. 28–54). Oxford University Press.
- Vaeth, M., Kahlfuss, S., & Feske, S. (2020). CRAC channels and calcium signaling in T cell-mediated immunity. Trends in Immunology , 41(10), 878–901. https://doi.org/10.1016/j.it.2020.06.012
- Zalk, R., Lehnart, S. E., & Marks, A. R. (2007). Modulation of the ryanodine receptor and intracellular calcium. Annual Review of Biochemistry , 76, 367–385. http://www.ncbi.nlm.nih.gov/pubmed/17506640
Internet Resources
Excitation and emission spectra of rhod-3.
Guideline for Generation of Stable Cell Lines. Technical Reference Guide. From Lonza. 2012.
A detailed description to generate stable cell lines.

