Ultra-deep ATAC-seq for sorted neurons
Andrea Kriz, Alisa Mo
Abstract
Isolation of neurons from frozen post-mortem human brain tissue and preparation of ATAC-seq libraries for ultra-deep sequencing and somatic mutation detection.
Steps
Isolation of Nuclei from Adult Human Brain Tissue (modified from Allen Human Brain Tissue PF0291)
Reagent list:
Buffer preparation: Buffer preparation: On the day prior to sorting, prepare the following buffers:
a. Nuclei Isolation Media (NIM) :
| A | B | C | D | E |
|---|---|---|---|---|
| Formula(in milliQ water) | Ingredients: | For 1.5 mL(1 ATAC-seq brain) | For 5 mL(3 ATAC-seq brains) | For 20 mL |
| 0.25M Sucrose | Sucrose | 0.13g | 0.43 g | 1.72 g |
| 25mM KCl | 2M KCl | 18.8 μL | 62.7 uL | 250.8 uL |
| 5mM MgCl2 | 1M MgCl2 | 7.5 uL | 25 uL | 100 uL |
| 10mM Tris-HCl, pH 8 | 1M Tris-HCl, pH 8 | 15 uL | 50 uL | 200 uL |
| 0.1% Triton X-100 | 10% Triton X-100 | 15 uL | 50 uL | 200 uL |
| Water | Water | ~1410 uL | ~4.7 mL | 18.8 mL |
Filter through 0.22µM filter and store at 4°C. Can keep for ~1-2 weeks.
b. Blocking buffer :
| A | B | C | D | E |
|---|---|---|---|---|
| Formula (in milliQ water) | Ingredients: | For 5 mL | For 10 mL | For 20 mL |
| Water | Water | 4.3 mL | 8.6 mL | 17.2 mL |
| 1X PBS | 10X PBS | 500 uL | 1000 uL | 2000 uL |
| 0.8% BSA | 20% BSA | 200 uL | 400 uL | 800 uL |
Store at 4°C. Can keep for ~1-2 weeks.
c. 1X Tagmentation buffer w/ BSA for ATAC-seq:
| A | B | C |
|---|---|---|
| Ingredients: | For 4 ATAC-seq reactions | For 6 ATAC-seq reactions |
| 2X Tagmentation buffer (Diagenode) | 220 uL | 330 uL |
| 5% digitonin | 0.88 uL | 1.3 uL |
| 10% Tween-20 | 4.4 uL | 6.6 uL |
| PBS | 145 uL | 218 uL |
| 20% BSA | 22 uL | 33 uL |
| Water | 25.5 uL | 38.3 uL |
Place at 4°C until ready to use.
Load Tn5: Mix 1 µL of annealed oligo A/oligo Rev with 1 µL of the annealed oligo B/oligo Rev as per Diagenode instructions.
Add 2 µL of Diagenode Tagmentase
Incubate at RT for 30 min
Add 2 µL glycerol. Place at -20°C until ready for use. Can store for a few week.
Tissue preparation:
Chill tweezers, scalpel, and dissecting plate (cell culture plate) on dry ice. Chill dounce homogenizers with B pestle on ice. Make sure swinging-bucket centrifuge is cooled to 4°C.
Spray down all surfaces and pipettes with 70% Ethanol and DNA Away
Make homogenization buffer (NIM + additives): Homogenization buffer: 5mL NIM + 5 uL 1M DTT (1 mM final concentration) + Mini Protease inhibitor (1/2 tablet) + 25 uL 100 mM spermidine (0.5 mM final concentration).
Scrape ~10mg frozen brain tissue onto cell culture plate, either on top of dry ice or in a cryostat at -20°C. Add 1.5 mL Homogenization buffer to tissue. Add tissue/buffer mix to Dounce homogenizer.
Dounce 15-20 strokes with B pestle. If the tissue is large, use more homogenization buffer (we have used up to 5 mL) and dounce with A pestle followed by B pestle.
Filter through 40µM filter into Eppendorf tube.
Spin in 4°C swinging-bucket centrifuge for 10min @ 900xg
While spin is going on, clean homogenizers by rinsing thoroughly with ddH2O, and then soaking them in 20% bleach for at least 20 minutes.
Make immunostaining buffer (Blocking buffer + 1:1000 dilution NeuN-488).
Remove supernatant. Do not disturb the pellet.
Add 1mL Blocking buffer + NeuN. Resuspend pellet gently with pipette. Rotate end-to-end in the cold room for 15-20 minutes.
Spin in 4°C swinging-bucket centrifuge for 5min @ 400xg
Make DAPI solution. First, dilute stock DAPI (1mg/mL) 1:15 in blocking buffer. Then, add 1 µL of the diluted dapi solution to 1 mL blocking buffer.
Remove supernatant from pellet.
Add 1mL Blocking buffer + Dapi and again resuspend. Strain through 40 µM filter into new Eppendorf tube immediately before nuclei sorting.
Prepare tubes for ATAC-seq: Add 60 µL of 1X Tagmentation buffer into each tube for ATAC-seq (2 tubes per brain)
ATAC-seq (modified from Omni-ATAC protocol (Corces et al., 2017))
Sort 10,000 NeuN+ nuclei into 60µL of tagmentation buffer per ATAC-seq (2 replicates per brain sample). Proceed immediately after sorting to ATAC-seq
After sorting, spin each tube in 4 deg swinging bucket centrifuge at 500xg for 5 minutes.
Remove ~75 µL of supernatant leaving ~10 µL at bottom. Tap tube gently to mix pellet.
Add 45 µL of 1X Tagmentation buffer. Do not mix.
Add 1 µL of loaded Tn5. Pipette gently 3-4 times and tap to mix.
Incubate at 37°C for 30 minutes in the Thermomixer without mixing. Gently tap tube at ~10 minutes and at ~20 minutes into the incubation to mix.
Stop tagmentation reaction by adding 250 µL of DNA binding buffer from Zymo Clean and Concentrator 5 kit. Add to sample tube, pipette 6x or vortex until homogenous.
Follow Zymo kit instructions to purify DNA:
- Add solution to spin-column tube. Spin at 10,000xg for 30 seconds.
- Wash x 200 µL wash buffer. Spin at 10,000xg for 30 seconds.
- Repeat wash x 200 µL wash buffer. Spin at 10,000xg for 30 seconds.
- Transfer spin-column tube into Eppendorf tube. Add 20 µL of water. Let sit for 1 minute. Then spin at 10,000xg for 1 minute.
Prepare PCR reaction on ice:
- 2.5 µL i5 primer
- 2.5 µL i7 primer
- 25 µL NEBNext High-Fidelity 2X PCR Master Mix, thaw on ice and invert until homogenous
- 20 µL purified tagmented DNA
PCR amplification:
- 72°C x 5 minutes
- 98°C x 30 seconds
- 98°C x 10 seconds
- 63°C x 30 seconds
- 72°C x 1 min
- Loop back to step [3] 8 times (9 cycles total)
- 72°C x 1 minute
- 4°C hold
Clean the library using Zymo clean and concentrator 5 kit. Add 250 µL of DNA binding buffer to PCR reaction and follow Zymo kit directions to purify.
Elute in 40-50 µL 0.1X TE (or Zymo elution buffer). Let sit for 1 minute, spin for 1 minute.
Run a 1:4 dilution of library on HS5000 Tapestation

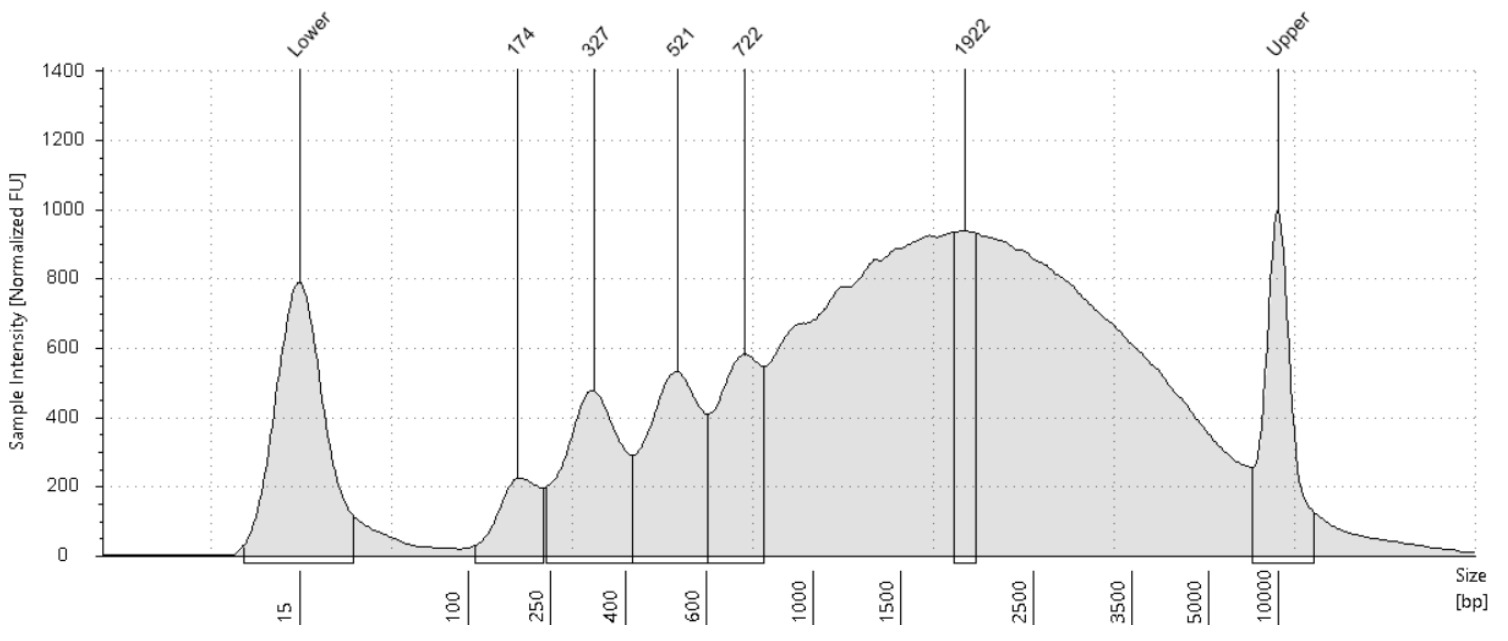
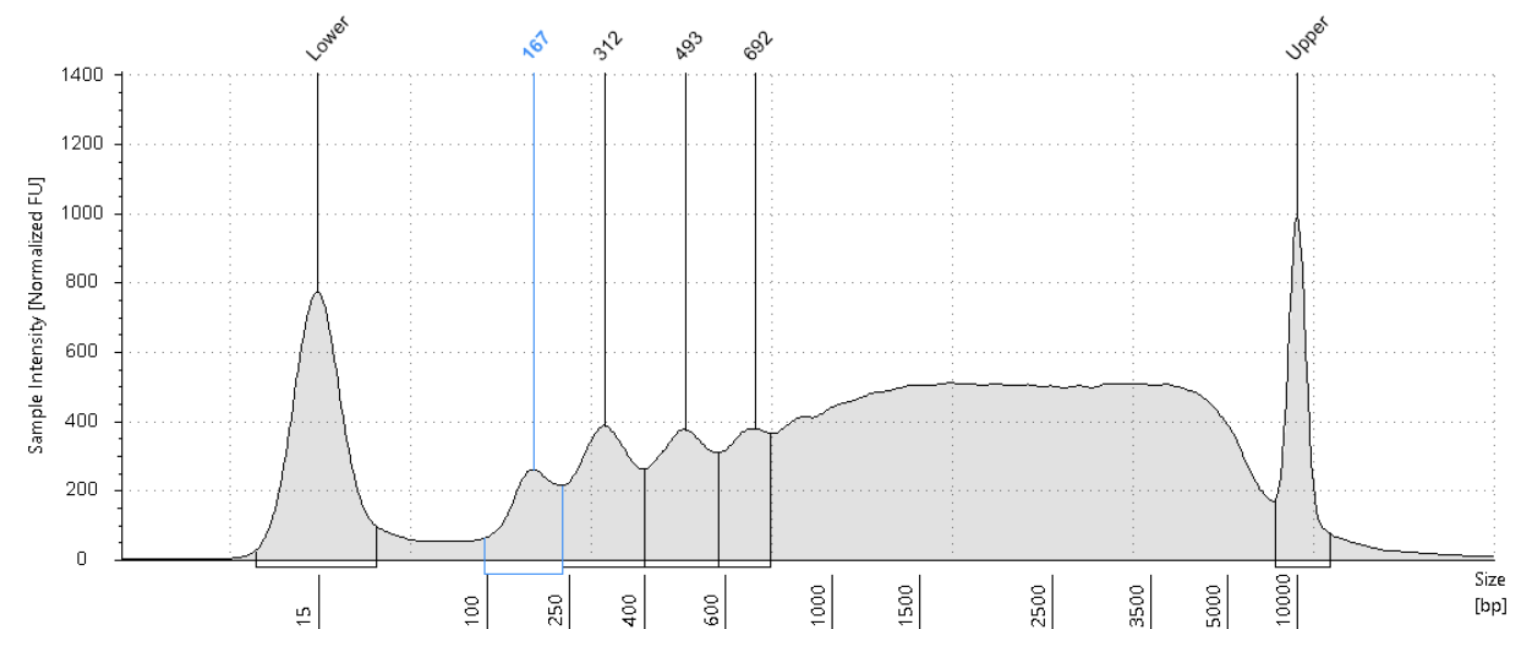
Store libraries at -20°C.

