Three-dimensional modeling of coral skeletal fragments for morphometrics
Colin J Anthony, Colin Lock, Bastian Bentlage, Renee Crisostomo
Abstract
This protocol details a comprehensive method for scanning and modeling coral fragments to accurately calculate surface area, volume, and other key morphological features essential for phenomics and ecophysiology studies. The procedure encompasses high-resolution 3D scanning, digital reconstruction, and data generation, thus providing precise quantification of coral morphologies. This protocol offers a standardized approach for morphological analyses, supporting diverse research applications in coral biology and ecology.
Steps
Skeletal Preparation
Airbrush tissue from coral skeleton with filtered seawater until only a bare skeleton remains.

Place fragments in drying oven overnight (50°C) to evaporate remaining water from fragment.
Weigh dried fragments.
This will be used later to calculate skeletal density.
Coat coral fragment in white, chalked spray paint.
We use RUST-OLEUM Chalked, Linen White spray paint (Internet # 307244837, Model # 339822). This reduces light scatter and allows the fragment to be detected by our 3D-scanner.
Depending on species, mount coral fragment on a metal pin to prevent scanner clip from covering up parts of the skeleton that are of interest.
For example, we mount staghorn Acropora branches on standard 26/6 staples with ULTRAGEL CONTROL Loctite super glue (#43903). This makes the model cleaning process much easier, as it provides a clear, easily patchable cutoff point for our coral model.
Scanning fragments and generating point clouds
Depending on your research question, there are two routes you can take to model your fragment: 1) simplified morphometric modeling or 2) corallite-focused morphometric modeling .
The simplified workflow (SW) uses less detailed models quickly calculate a few informative metrics, such as skeletal density or complexity. The simplified workflow is also often paired with eco-physiological workflows, such as Symbiodiniaceae cell counting, as it inherently calculates skeletal surface area, which can be used to normalize cell counts. The corallite-focused workflow (CW) allows you to quantify characteristics traditionally used in taxonomy, but also requires more computing power and time to complete. Also note, the corallite-focused workflow can generate all of the same characters as those described in the simplified workflow.
Critical steps indicate steps that differ between the two workflows. Within these steps, differences will be indicated by wither SW or CW .
| A | B |
|---|---|
| Skeletal surface area & volume | Only used to normalize other measurements |
| Skeletal density | Ratio of fragment weight to volume |
| Skeletal complexity | Ratio of fragment area to volume |
| Corallite density | Number of non-emersed corallites per cm^2 |
Table 1. Simple characters measured in simplified morphometric modeling
| A | B |
|---|---|
| Radial crowding | Average number of emersed radial corallites /3 transects |
| Radial wall thickness | Width of radial wall |
| Radial corallite profile length | Maximum distance from base to outer edge of radial corallite |
| Radial corallite diameter | Maximum diameter of radial corallite from inner to outer wall |
| Radial calice diameter | Maximum diameter of radial calice from inner to outer wall |
| Axial tip diameter | Maximum diameter of axial tip |
Table 2. Traditional taxonomic characters (Wallace, 1999) measured in corallite-focused modeling
Mount fragment on 3D scanner. We use a D3D-s jewelry scanner, but many scanners can be used, with local protocol modification.
Name project and set directory for point cloud outputs.
Set the scanning length and diameter to ~1.5x the length and diameter of the coral fragment.
This accounts for variation, and ensures portions of the fragment are not missed during the scanning process.
Set the scanning depth to 1.3-1.5x the diameter used in the previous step.
Set horizontal turns, verticle turns, and/or horizontal angles to ones that sufficiently capture your coral.
For Acropora fragments (20x30mm), we do 12 vertical turns , and scan from 0 and 70 degrees (SW) or 4 horizontal turns and 12 vertical turns (CW) . This will require local modification and fine tuning depending on coral morphology and fragment size. The goal is to sufficiently scan the entire surface area in a minimal amount of time. Generally, more turns produce a better model
SW - Set resolution to medium or high , depending on your coral's corallite complexity.
CW - Set resolution to maximum
Set camera gain to 3. (May be adjusted locally).
Start scanning.
Modeling in MeshLab
Align point clouds
Click the "Align" tool (Yellow circle with black "A" on the top tool bar) > "Glue Here Visible Meshes" > edit default ICP parameters to sample 6000 points (CW) > "Process" > Close the "Align Tool" window
I like to make sure my Avg Err is below 0.04.
Reconstruct fragment surface
"Filters" > "Remeshing, Simplification and Reconstruction" > "Surface Reconstruction: Screened Poisson" > Check box next to "Merge all visible layers" > Set reconstruction depth to 9 (SW) or 11 (CW) > Check box next to "Pre-Clean" > "Apply" > Close "Surface Reconstruction: Screened Poisson" window > Hold "Ctrl" and click on the eyeball next to "Poisson mesh" on the right side of the screen > right click "Poisson mesh" > "Duplicate Current layer" (MeshLab does not have an undo function, so duplicating your mesh serves as a good backup) > Close the eyeball next to the non-selected Poisson mesh
Remove mount from surface reconstruction
"Select Faces in a rectangular Region" > highlight and delete all portions of the reconstruction that are not part of the coral by dragging the tool over unwanted regions and pressing "delete"
Close holes in reconstruction
"Filters" > "Remeshing, Simplification and Reconstruction" > "Close Holes" > set "Max size to be closed" to 5000 > "Apply"
Sometimes, it doesn't work properly, so you may need to redo or revisit step 18 with mesh copy generated in step 17 until it can effectively close the hole.
Calculate skeletal volume
"Filters" > "Quality Measure and Computations" > "Compute Geometric Measures" > Record "Mesh Volume"
Calculate skeletal surface area
If normalizing cell counts or corallites to skeletal surface area, delete all faces that did not contain live tissue (see Step 18). Recalculate surface area (see Step 20).
Calculate corallite density (CS) and radial crowding (CW)
Select "Z-painting" paint brush > Simultaneously use a Portable Incremental Counter and the paint brush tool to count non-emersed corallites for corallite density (CS) and emersed corallites for radial crowding > Record number of corallites, normalize to surface area calculated in step 21.
Emersed corallites require a higher resolution model for consistent detection, hence their inclusion in the CW workflow and not the CS workflow.
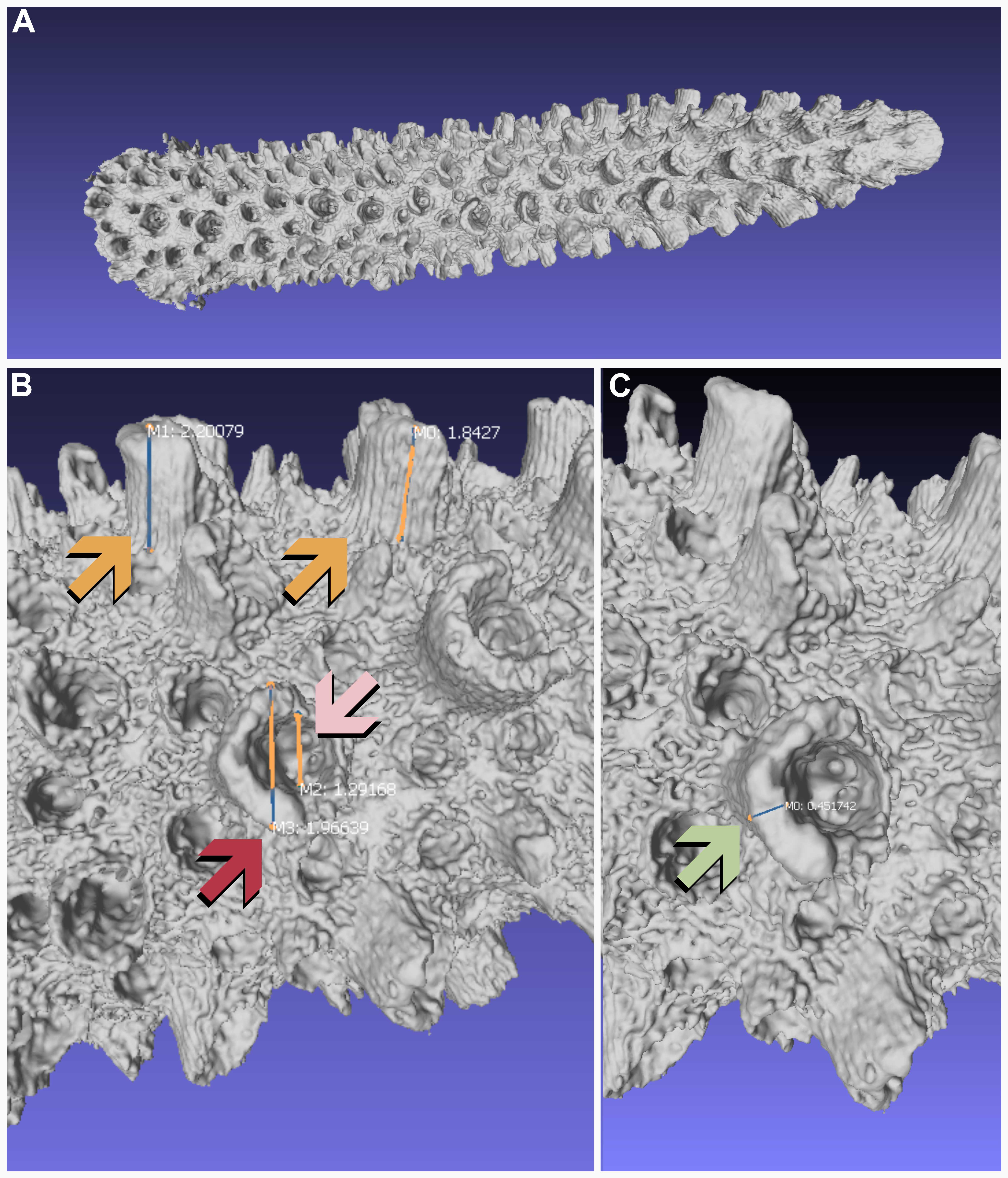
Calculate radial wall thickness, radial corallite profile length, radial corallite diameter, radial calice diameter, and axial tip diameter (CW) using the built-in "Measuring Tool" within MeshLab.
For Acropora fragments, we collect a subsample of ~30 corallites per fragment per character. Radial wall thickness, radial corallite profile length, radial corallite diameter, and radial calice diameter measurements.
The first 2 cm of each fragment were not sampled to ensure that measurements of immature corallites did not skew measurements of matured corallites. Axial tip diameter only produced one measurement per fragment.
Export final Mesh and close MeshLab
File > "Export Mesh As..." > Name file > "Save" > Close MeshLab


