Slide-TCR-Seq v3 (IVT)
Fei Chen, Sophia Liu, Ruth Raichur
T cell receptor
spatial transcriptomics
tertiary lymphoid structures
cancer niches
Slide-TCR-seq
IVT
in vitro transcription
Abstract
T cells mediate antigen-specific immune responses to disease through the specificity and diversity of their clonotypic T cell receptors (TCRs). Determining the spatial distributions of T cell clonotypes in tissues is essential to understanding T cell behavior, but spatial sequencing methods remain unable to profile the TCR repertoire.
We developed Slide-TCR-seq, a 10-μm-resolution method, to sequence whole transcriptomes and TCRs within intact tissues. Our method yields insights into the spatial relationships between clonality, neighboring cell types, and gene expression that drive T cell responses.
The most recent version of our protocol uses in vitro transcription in lieu of rhPCR amplification, which overcomes the barcode switching introduced by the rhPCR and results in higher mapping rates.
Steps
PCR to add T7 to cDNA libraries
This protocol amplifies TCRs from unfragmented, full-length cDNA from Slide-Seq.
Prepare two 10-nanogram* dilutions of all samples into 12.25 μL of ultrapure water for amplifying TCR alpha and beta sequences in separate reactions.
*We have successfully tested down to 2 ng for low-concentration samples.
Prepare the primer extension PCR master mix using KAPA Hifi Hotstart Readymix 2X and T7-TCRV primer pool. Gently mix by pipetting and run the PCR program below.
Note: TRAV and TRBV primers are pooled separately and are treated as individual reactions for each sample.
Primer extension PCR mix per sample:
| A | B | C |
|---|---|---|
| Volume (μL) | Reagent | Final concentration |
| 12.5 | KAPA Hifi Hotstart Readymix 2X | 1 X |
| 0.25 | 100 μM T7-TCRV primer pool | 1 μM |
| 4-10 ng | cDNA | |
| up to 25 μL | Ultrapure water |
Final volume 25 μL
Primer extension PCR protocol:
| A | B | C |
|---|---|---|
| Cycles | Temp | Time |
| 5 | 95 °C | 5 minutes |
| 65 °C for human primer pool/70 °C for mouse primer pool | 30 seconds | |
| 72 °C | 3 minutes | |
| 1 | 4 °C | hold |
Safe stopping point, store at 4 °C
Add 25 μL of water to bring the reaction to 50 μL. Perform a PCR clean-up following the manufacturer's instructions using SPRIselect or AMPure XP beads at 0.6X (30 μL of SPRI beads to 50 μLPCR reaction volume). Elute in 9 μL of water.
Prepare the T7/Truseq PCR master mix with KAPA Hifi Hotstart Readymix 2X. Add 16 μL of master mix to 9 μL of the sample. Gently mix by pipetting and run the PCR program below.
T7/Truseq PCR mix per sample:
| A | B | C |
|---|---|---|
| Volume (uL) | Reagent | Final concentration |
| 12.5 | KAPA Hifi Hotstart Readymix 2X | 1 X |
| 0.5 | 100uM Truseq-P5 Hybrid primer | 2 μM |
| 0.5 | 100uM T7 PCR primer | 2 μM |
| 9 | Sample | |
| 2.5 | Ultrapure water (up to 25) |
Final volume 25 μL
T7/Truseq PCR protocol:
| A | B | C |
|---|---|---|
| Cycles | Temp | Time |
| 1 | 98 °C | 2 minutes |
| 10 | 98 °C | 1 minute |
| 60 °C | 30 seconds | |
| 72 °C | 3 minutes | |
| 1 | 72 °C | 5 minutes |
| 4 °C | hold |
Safe stopping point, store at 4 °C
Add 25 μL of water to bring the reaction to 50 μL. Perform a PCR clean-up following the manufacturer's instructions using SPRIselect or AMPure XP beads at 0.6X (30 μL of SPRI beads to 50 μLPCR reaction volume). Elute in 8 μL of water.
IVT amplification
Follow the manufacturer's instructions on the HiScribe RNA synthesis kit using 8 μL of the sample eluted in the previous step. Incubate reaction for 2 hours at 37 °C.
HiScribe RNA Synthesis mix per sample:
| A | B | C |
|---|---|---|
| Volume (uL) | Reagent | Final concentration |
| 10 | NTP Buffer Mix | 10 mM each NTP |
| 2 | T7 RNA Polymerase Mix | |
| 8 | Sample |
Final volume 20 μL
Use RNAse away to clean all surfaces and pipettors.
Add 30 μL of water to bring the reaction to 50 μL. Perform a PCR clean-up using SPRIselect or AMPure XP beads at 0.6X (30 μL of SPRI beads to 50 μL PCR reaction volume), following the steps below:
For a 50 μL reaction, add 30 uL of SPRI beads.
Incubate for 5 minutes at RT.
Incubate for 2 minutes on a magnet until the solution turns clear.
Discard supernatant.
Wash on a magnet for 30 sec with 200 μL of freshly made 80% EtOH.
Repeat wash.
Discard supernatant.
Spin down briefly on a table spinner.
Remove all EtOH with a 20 μL pipette.
Elute with 20 μL of H2O.
Use a NanoDrop on the RNA setting to measure RNA concentration.
RT
Add 180 μL of the following RT mix to 20 μL of the RNA sample. Incubate reaction for 2 hours at 42 °C.
Reverse Transcription Mix per sample:
| A | B | C |
|---|---|---|
| Volume (uL) | Reagent | Final concentration |
| 40 | Maxima 5X RT buffer | 1 X |
| 20 | 10 mM dNTPs | 1 mM |
| 5 | RNAse inhibitor | |
| 2 | 100 μM Truseq-P5 Hybrid primer | 1 μM |
| 10 | Maxima H-RTase | |
| 20 | Template RNA (sample) | 1 pg - 5 ug |
| 103 | Ultrapure water (up to 200) |
Final volume 200 μL
Safe stopping point, store at 4 °C
Perform a PCR clean-up following the manufacturer's instructions using SPRIselect or AMPure XP beads at 0.6X (120 μL of SPRI beads to 200 μL RT reaction volume). Elute in 20 μL of water. Record concentrations using a NanoDrop on the ssDNA setting and save all samples.
Index PCR
Prepare the PCR master mix with KAPA Hifi Hotstart Readymix 2X, P5-Truseq PCR primer, and Nextera PCR primer index. Gently mix by pipetting and divide the total volume of each sample into 4 PCR tubes each containing 50 μL (25%) of the total.
Note: Each sample must use a different i7 index if you intend to pool samples for multiplexed sequencing. We do not recommend dual-indexing of samples.
Index PCR mix per sample:
| A | B | C |
|---|---|---|
| Volume (uL) | Reagent | Final Concentration |
| 100 | KAPA Hifi Hotstart Readymix 2X | 1 X |
| 4 | 100 μM Truseq-P5 Hybrid PCR primer | 2 μM |
| 4 | 100 μM Nextera PCR primer (i7) | 2 μM |
| 100 ng | Sample | |
| up to 200 | ultrapure water |
Final volume 200 μL
Index PCR protocol:
| A | B | C |
|---|---|---|
| Cycles | Temp | Time |
| 1 | 98 °C | 2 minutes |
| 10 | 98 °C | 1 minute |
| 67 °C | 20 seconds | |
| 72 °C | 3 minutes | |
| 1 | 72 °C | 5 minutes |
| 4 °C | hold |
Safe stopping point, store at 4 °C
Recombine the samples that were split into 4 parts in the previous step and perform PCR clean-up following the manufacturer's instructions using SPRIselect or AMPure XP beads at 0.6X (120 μL of SPRI beads to 200 μL PCR reaction volume). Elute in 10 μL of water.
To quantify the TCR libraries, use the Qubit dsDNA high-sensitivity kit and BioAnalyzer High-SensitivityThe expected DNA kit following the manufacturer protocols.
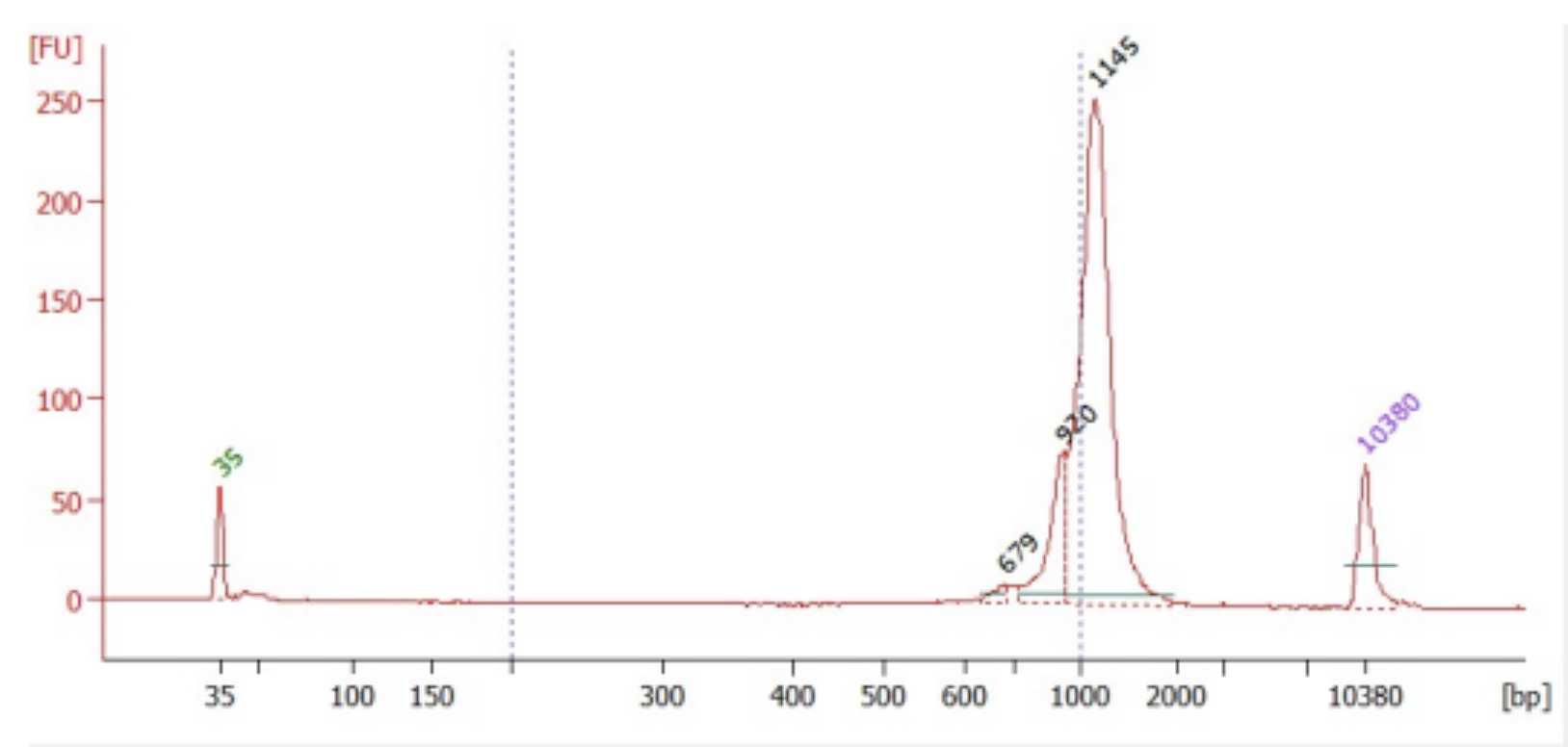
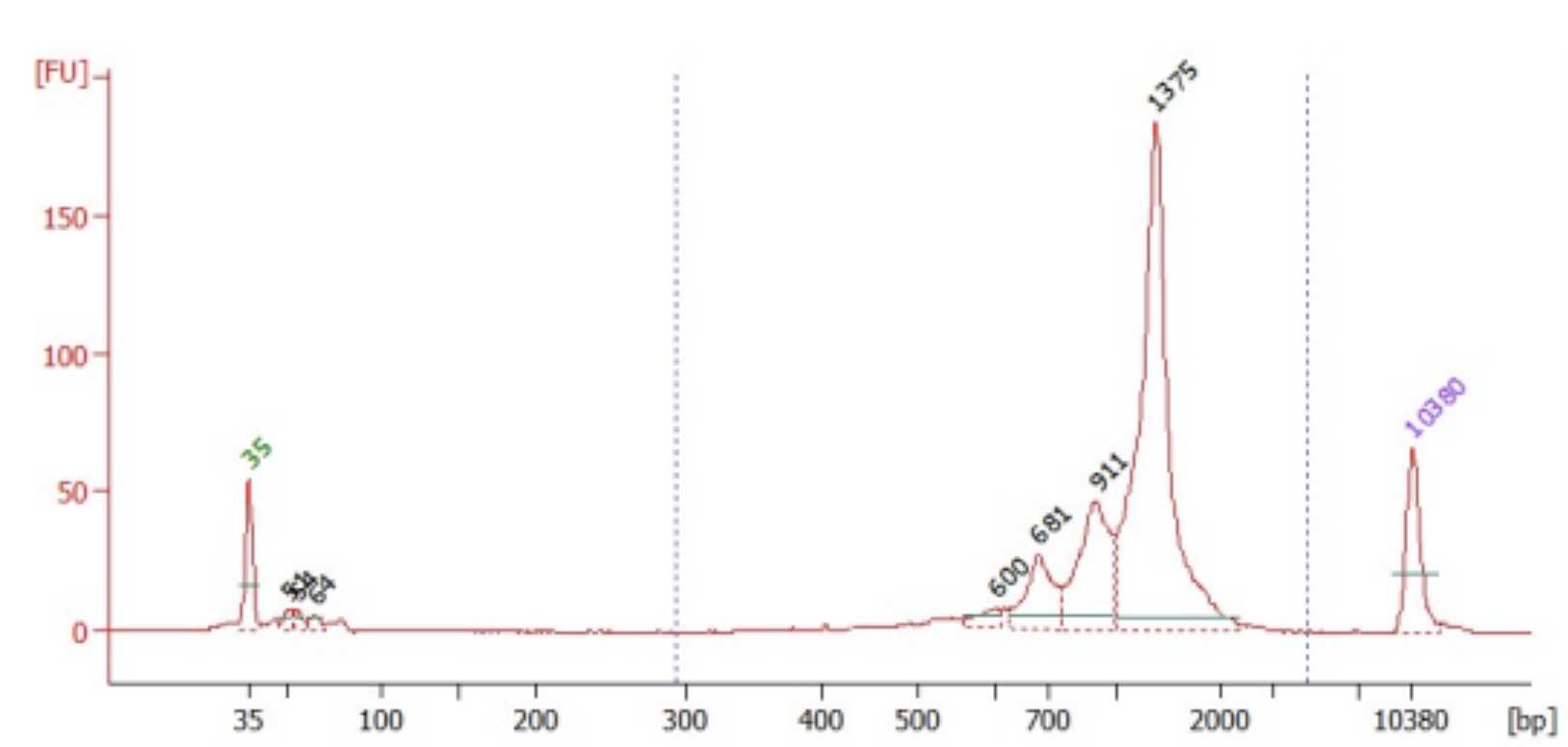
Sequencing
TRA libraries are best sequenced on a Nanopore. TRB libraries can be sequenced on a Nanopore or MiSeq.
For best results, it's generally advised to sequence each sample to a depth of 1-2 million reads.
MiSeq read structure is as follows:
Read 1: 42 bp
Index 1: 8 bp
Read 2: 270 bp
Index 2: 0 bp

