Single coacervate sequencing
Franziska Aron, Damian Wollny
Disclaimer
DISCLAIMER – FOR INFORMATIONAL PURPOSES ONLY; USE AT YOUR OWN RISK
The protocol content here is for informational purposes only and does not constitute legal, medical, clinical, or safety advice, or otherwise; content added to protocols.io is not peer reviewed and may not have undergone a formal approval of any kind. Information presented in this protocol should not substitute for independent professional judgment, advice, diagnosis, or treatment. Any action you take or refrain from taking using or relying upon the information presented here is strictly at your own risk. You agree that neither the Company nor any of the authors, contributors, administrators, or anyone else associated with protocols.io, can be held responsible for your use of the information contained in or linked to this protocol or any of our Sites/Apps and Services.
Abstract
Here, we present a protocol which enables the comprehensive characterization of the RNA content of single phase-separated coacervates. We adapted single-cell RNA sequencing technology in combination with fluorescence activated cell sorting (FACS) to answer the question of how one condensate differs from the other in terms of RNA composition and how it relates to condensate features such as droplet size. This approach represents a powerful addition to labor intensive and low throughput microscopy approaches which have been the state of the art approach for coacervate RNA characterization. This protocol includes droplet production, as well as a Smart-seq2 protocol adaption for lysis, reverse transcription and cDNA amplification and sequencing library preparation. This protocol ends with the library preparation. Afterwards it got sequenced on an Illumina NextSeq500 (paired end for 300 cycles).
The Smart-seq2 protocol was originally published in Picelli, S., Faridani, O., Björklund, Å. et al. Full-length RNA-seq from single cells using Smart-seq2. Nat Protoc 9, 171–181 (2014). https://doi.org/10.1038/nprot.2014.006
Before start
-
Take the Agencourt RNAClean XP beads out of the fridge and let them adjust to room temperature (=RT), aliquot 110µl in each tube of a 8-PCR-stripe, vortex in between to keep the beads in solution
-
Defrost the reagents for the Master Mix I and Master Mix II (except enzymes and TSO)
-
Unpack RNase free filter tips (10x of 10µl filter tips; 10x 200µl filter tips; 5x 20µl filter tips)
-
Prepare fresh 80% EtOH p.a. (= 50ml per plate)
-
Get a box full of ice
This protocol is designed for a 96-well plate (LoBind).
Steps
Buffer preparation
Prepare 6Molarity (M) Guanidinium hydrochloride
Prepare the droplet buffer consisting of 10millimolar (mM)Tris and 4millimolar (mM)MgCl2 8
Droplet production
Calculate the droplet production according to your experiment.
The example is made for CM-Dextran:PDDA coacervates (molar ratio: 6:1)
| A | B | C | D | E |
|---|---|---|---|---|
| 1000µl Droplets | Stock concentration | Final concentration | ||
| Reagents | g/mol | M | mM | µl use |
| CM-Dextran Sodium Salt | 162.14 | 1 | 60 | 60 |
| PDDA | 174 | 1 | 10 | 5.8 |
| ng/µl | ng/µl | µl use | ||
| RNA | 1243 | 50 | 40.2 | |
| Droplet Buffer | 10mM Tris, 4mM MgCl2, pH 8 | 893.7 |
Calculate how much RNA you need to add to the entire solution for a final concentration of50ng/µl
Mix the droplet buffer with the CM-Dextran Sodium Salt
Add the RNA and mix briefly
Finally add the PDDA [=Poly-{diallyl-dimethylammoniumchlorid}-solution] and mix by vortexing
Add 4µl of 6Molarity (M) Guanidinium hydrochloride into each well of the full skirted 96-well LoBind plate
Perform a droplet sorting via FACS into the 96-well plate. Store the sorted droplets immediately at -80°Cor directly continue with the protocol.
Smart-seq2 preparation
Prepare Master Mix I
| A | B | C |
|---|---|---|
| Reagents | Stock concentration | 1x [µl] |
| oligo-dT primer | 10 µM | 1 |
| dNTPs | 10 mM each | 1 |
| nuclease free H2O | 2 | |
| MasterMix Total [µl] | 4 | |
| Assay Total [µl] | 4 |
add all the reagents, mix by flicking the tube and spin down
Prepare Master Mix II
| A | B | C |
|---|---|---|
| Reagents | Stock concentration | 1x [µl] |
| nuclease free H2O | 0.09 | |
| MgCl2 | 1 M | 0.06 |
| Betaine | 5 M | 2 |
| DTT | 100 mM | 0.5 |
| 5xI FS Buffer | 5x | 2 |
| RNAse inhibitor | 40 U/µl | 0.25 |
| Superscript II RT | 200 U/µl | 0.5 |
| TSO | 100 µM | 0.1 |
| MM total [µl] | 5.5 | |
| Assay total [µl] | 9.5 |
add all the reagents, mix by flicking the tube and spin down
Droplet clean up
Mix the amount of droplets with RNA XP beads in a 1:2.2 ratio
Vortex and spin down briefly
Incubate for 0h 5m 0s at 72Room temperature
Pellet the beads on a magnet until solution is clear (~0h 5m 0s)
Discard supernatant
Add 180µL of 80% EtOH to the bead pellet
Discard EtOH
Add 160µL of 80% EtOH to the bead pellet
Discard EtOH
Remove the EtOH completely
Resuspend the pellet in 4µL of Master Mix I by pipetting up and down
(The plate is not on the magnet for elution)
Smart-seq2
Incubate at 72°C for0h 3m 0s
Spin down the reaction tube
Add 5.5µL of Master Mix II, mix by flicking the tube, spin down and start the following incubation
| A | B | C |
|---|---|---|
| Cycle [Amount] | Temp [°C] | Time [min] |
| 1 | 42 | 90 |
| 10 | 50 | 2 |
| 42 | 2 | |
| 1 | 70 | 15 |
| 4 | Hold |
Prepare Master Mix III
| A | B | C |
|---|---|---|
| Reagents | Stock concentration | 1x [µl] |
| KAPA HiFi HS ReadyMix | 2 x | 12.5 |
| IS PCR primer | 10 µM | 0.25 |
| nuclease free H2O | 2.25 | |
| MM total [µl] | 15 | |
| Assay total [µl] | 24.5 |
Mix by flicking the tube and spin down
Add 15µL of Master Mix III to the reaction
Mix by flicking the tube and spin down
Start the following incubation
| A | B | C |
|---|---|---|
| Cycle [Amount] | Temp [°C] | Time |
| 1 | 98 | 3 min |
| 12 - 23 | 98 | 20 sec |
| 67 | 15 sec | |
| 72 | 6 min | |
| 1 | 72 | 5 min |
| 4 | Hold |
Add SPRIselect beads in a 1:0.7 ratio, vortex and spin down briefly
Incubate for 0h 5m 0s at Room temperature
Pellet the beads on a magnet until solution is clear (~0h 5m 0s)
Discard supernatant
Add 180µL of 80% EtOH (= Ethanol, molecular biology grade) to the bead pellet
Discard EtOH
Add 160µL of 80% EtOH to the bead pellet
Discard EtOH
Remove the EtOH completely
Resuspend the pellet in 17.5µL nuclease free H2O by pipetting up and down
(The plate is not on the magnet for elution)
Transfer the eluate into a fresh 96-well plate
Store at -20°Cuntil further usage
First Quality control
Quality Control
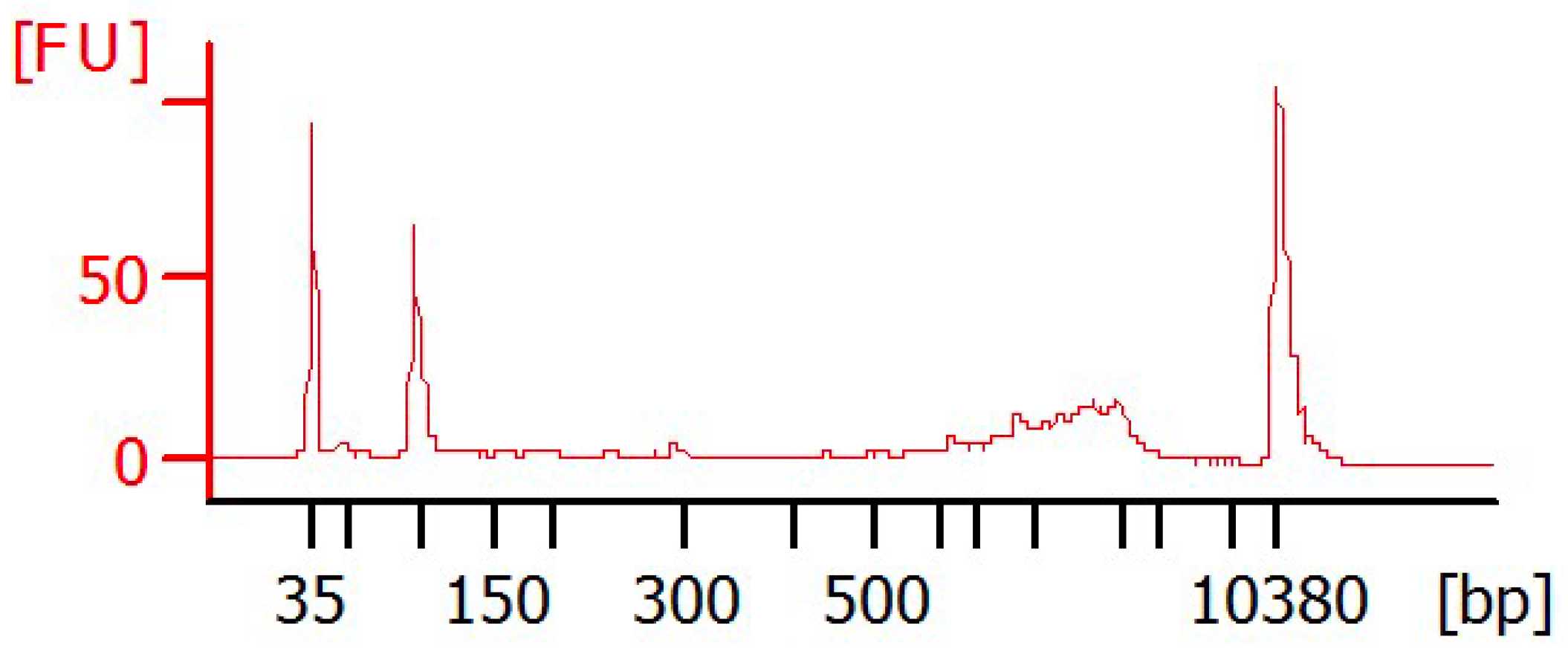
First measure 1µL of cDNA with the Qubit HS DNA Kit according to the manufacturer's protocol
Tagmentation
Get a box full of ice, bring Tagment DNA buffer and NT buffer (Illumina, Nextera XT Library Prep Kit) to room temperature
Decide already about the index combinations that you want to use
Predilute cDNA to a final concentration of 0.1ng/µl to 0.3ng/µl in nuclease free H2O
Prepare tagmentation pre-mix as described in the following table
| A | B |
|---|---|
| Reagent | 1x (µL) |
| Tagment DNA buffer | 2.5 |
| Amplicon Tagmentation mix (Tn5) | 1.25 |
| MM total (µL) | 3.75 |
Mix by vortexing and spin down briefly
Mix tagmentation pre-mix with pre-diluted cDNA as described following
| A | B |
|---|---|
| Reagent | 1x (µL) |
| tagmentation pre-mix | 3.75 |
| pre-diluted cDNA | 1.25 |
| Assay total [µl] | 5 |
Incubate in a thermocycler as described following
| A | B | C |
|---|---|---|
| Cycle [Amount] | Temp [°C] | Time [min] |
| 1 | 55 | 10 |
| 1 | 10 | Hold |
Spin down briefly
Add 1.25µL NT buffer to each reaction,
Mix by vortexing and spin down briefly
Tagmentation_Indexing
Prepare the indexing reaction as described in the following table and add to the reaction
| A | B |
|---|---|
| Reagents | 1x (µL) |
| Index primer 1 | 1.25 |
| Index primer 2 | 1.25 |
| Nextera PCR Master mix | 3.75 |
| MM total [µl] | 6.25 |
| Assay total [µl] | 12.5 |
Mix by flicking the tube and spin down
Start incubation in a thermocycler as described following
| A | B | C |
|---|---|---|
| Cycle [Amount] | Temp [°C] | Time |
| 1 | 72 | 3 min |
| 1 | 95 | 30 sec |
| 12 | 95 | 10 sec |
| 55 | 30 sec | |
| 72 | 60 sec | |
| 1 | 72 | 5 min |
| 1 | 10 | hold |
First bead clean up
Pool all the samples
Add SPRIselect beads in a 1:1 ratio, mix by vortexing
Incubate on a rotator for0h 5m 0s at Room temperature
Pellet the beads on a magnet until the solution is clear (~0h 5m 0s )
Discard the supernatant
Wash pellet with fresh 80% EtOH p.a.
Discard the supernatant
Repeat for a total of two washes
Remove the remaining EtOH
Wait0h 0m 30s to 0h 1m 0s
Elute in 1/5 of the sample volume in nuclease free H2O
Incubate for 0h 5m 0s at Room temperature
(The incubation tube is not on magnet for elution)
Incubate on a magnet until the solution is clear (~0h 5m 0s )
Second bead clean up
Transfer the supernatant into the prepared SPRIselect beads (1:0.8), mix by vortexing
Incubate on a rotator for0h 5m 0s at -20Room temperature
Pellet the beads on a magnet until the solution is clear (~0h 5m 0s )
Discard the supernatant
Wash pellet with fresh 80% EtOH
Discard the supernatant
Repeat for a total of two washes
Remove the reminaing EtOH
Wait 0h 0m 30s to 0h 1m 0s
Elute in 1/5 of the sample volume in nuclease free H2O
Incubate for 0h 5m 0s at-20Room temperature
Incuabte on a magnet until the solution is clear (~0h 5m 0s )
Transfer the clear supernatant to a fresh tube
Store at-20°Cuntil further usage
Second Quality control
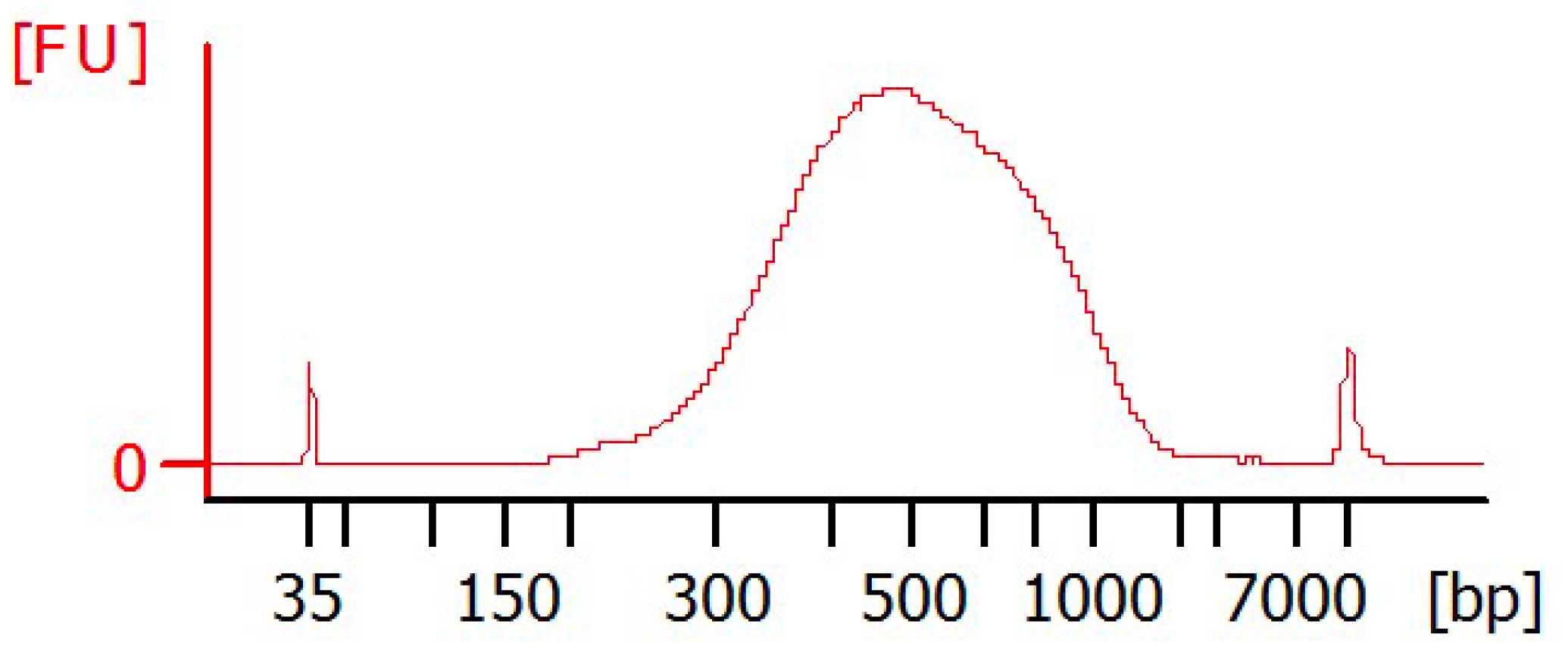
Quality control
First measure 1µL of cDNA with the Qubit HS ds Kit according to the manufacturer's protocol