Semi-automated protocol to quantify and characterize fluorescent three-dimensional vascular images
Chuo Fang, David H. Cribbs, Danny F Xie, Christian Crouzet, Krystal LoPresti, Yuke Wang, Christopher Robinson, William Jones, Fjolla Muqolli, Mark Fisher, Bernard Choi
Abstract
The microvasculature facilitates gas exchange, provides nutrients to cells, and regulates blood flow in response to stimuli. Vascular abnormalities are an indicator of pathology for various conditions, such as compromised vessel integrity in small vessel disease and angiogenesis in tumors. Traditional immunohistochemistry enables visualization of tissue cross-sections containing exogenously labeled vasculature. Although this approach can be utilized to quantify vascular changes within small fields-of-view, it is not a practical way to study the vasculature on the scale of whole organs. Three-dimensional (3D) imaging presents a more appropriate method to visualize the vascular architecture in tissue. Here we describe the complete protocol that we use to characterize the vasculature of different organs in mice encompassing the methods to fluorescently label vessels, optically clear tissue, collect 3D vascular images, and quantify these vascular images with a semi-automated approach. Applying this procedure of image analysis presents a method to reliably quantify and characterize vascular networks in a timely fashion. This procedure is also applicable to other methods of tissue clearing and vascular labels that generate 3D images of microvasculature.
Steps
Cardiac perfusion and retrooribtal injection
Cardiac perfusion
Begin by anesthetizing a mouse using an isoflurane chamber with 1.5 L/min of oxygen and 4.0% isoflurane.
Perfuse 10 mL of formalin at a rate of 2 mL/min
Once the mouse is anesthetized, remove it from the chamber and place its snout in a nose cone at 1.5 L/min oxygen and 1.5% isoflurane.
Administer a solution of lectin-DyLight-649 (200 µL, 25% lectin-DyLight and 75% saline) via retroorbital injection.
Allow the solution to circulate for about 20 minutes before proceeding with the cardiac perfusion.
It is recommended to perform the cardiac perfusion on a surgical tray or similar platform to contain the exsanguinated blood. Confirm that the mouse is at an appropriate plane of anesthesia using toe and/or tail pinches.
Next, open the chest cavity by performing a horizontal incision beneath the rib cage and a vertical incision along both sides of the chest.
.
Use hemostats to assist with holding the chest open to access the heart.
Perform a small incision on the right atrium of the heart to allow blood to exit the body.
Insert a butterfly needle into the left ventricle of the heart. Use a syringe pump to perfuse 10 mL of saline into the heart at a rate of 2 mL/min.
Brain extraction
Brain extraction
Remove the head by cutting caudal of the skull with scissors (bone scissors or large scissors preferred).
Cut the scalp to create two folds and expose the cranium.
Using angled microscissors, gently cut upwards from the foramen magnum to approximately the location of bregma on the brain.
Using a set of scissors, perform a lateral cut directly rostral to the olfactory bulbs. This cut should split the remainder of the skull down the centerline beyond bregma.
Use fine tweezers or a spatula to pry open each hemisphere of the skull.
Gently separate the brain from the base of the skull with a spatula. Sever any nerves connecting the brain to the skull.
Place the brain in ~10 mL of 10% formalin to completely submerge the brain. Store away from light.
After 24h, store the brain in PBS with 0.02% sodium azide at 4°C until further tissue processing. Store away from light.
(Optional) Exogenous labeling of hemosiderin with Prussian blue for cerebral microhemorrhage visualization.
The following procedures are based on 1-mm thick coronal sections of a bisected brain. Volumes and times may need to be adjusted for tissues of different sizes.
Prepare a working solution of 10% w/v of potassium ferrocyanide (10 g of potassium ferrocyanide per 100 mL of DIW).
Use a magnetic stir plate to mix the solution for at least 20 minutes.
Prepare a working solution of hydrochloric acid that is 20% of stock hydrochloric acid.
Mix the solutions of potassium ferrocyanide and hydrochloric acid in a 1:1 ratio. Approximately 5 mL of the mixed solution is used per sample. This solution should be prepared prior to each staining session.
Wash samples in 5 mL of DIW with shaking.
Submerge each sample into 5 mL of the working potassium ferrocyanide/hydrochloric acid solution for 1 hour
Perform a final DIW wash for 5 minutes.
Store the samples in PBS with 0.02% sodium azide at 4°C and away from light.
Tissue clearing
The following procedure is modified from the established iDISCO protocols. Wash durations were modified for 1-mm thick coronal sections of a bisected brain.
Perform a series of methanol washes (20%, 40%, 60%, 80%, 100%, and 100%, balance DIW) each for 20 minutes with shaking. Microcentrifuge tubes of 1.5 mL (or larger for larger samples) are recommended. Fill the tubes fully to minimize exposure to oxygen.
Incubate the samples in a solution of 66% dichloromethane and 33% methanol for one hour with shaking. The sample may be stored overnight in this situation if desired, without shaking.
Incubate the sample in dichloromethane twice for 15 minutes with shaking.
Store the samples in dibenzyl ether at 4°C until imaging.
Imaging with confocal microscopy
When imaging samples that have been cleared following the iDISCO protocol, it is recommended to image the sample while submerged in dibenzyl ether. There are various methods to safely house a sample with dibenzyl ether and protect the imaging objective. A straightforward method is to create an epoxy well on a cover glass to surround the sample and hold the dibenzyl ether.
To image the vasculature labeled with the lectin-DyLight-649, use an appropriate wavelength (633 nm can work) with an emission band of approximately 650-750 nm.
To visualize microhemorrhages labeled with Prussian blue, locate regions of Prussian blue positivity using a white light source.
Simultaneously collect a fluorescent image of the vasculature and a transmittance image to co-register the vascular fluorescence with cerebral microhemorrhages. This workflow along with representative images are depicted in Fig 1 .
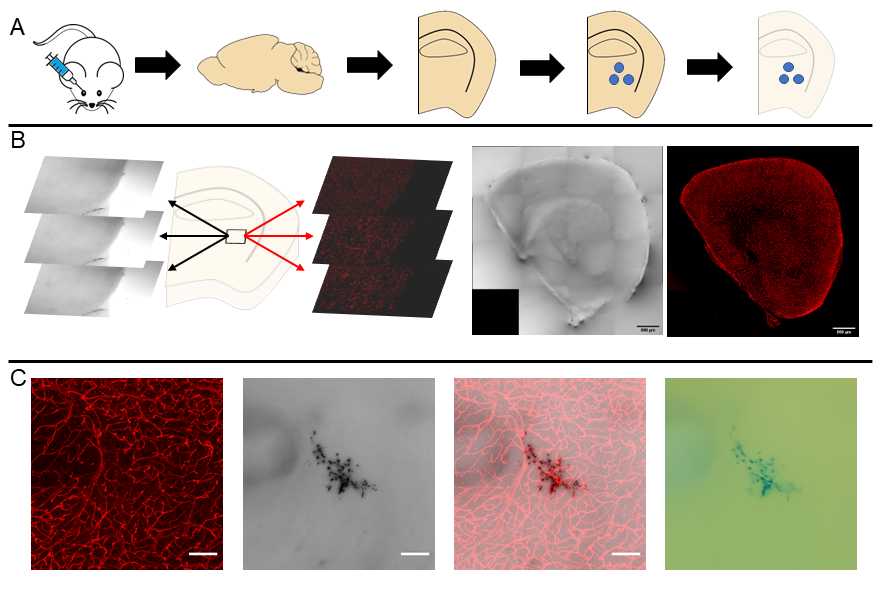
Our presented results are primarily from brain samples, but the procedures can be easily translated to other organs. Light-sheet microscopy can be used as an alternative to confocal microscopy to rapidly generate 3D reconstructions of the vascular network with minimal photobleaching.
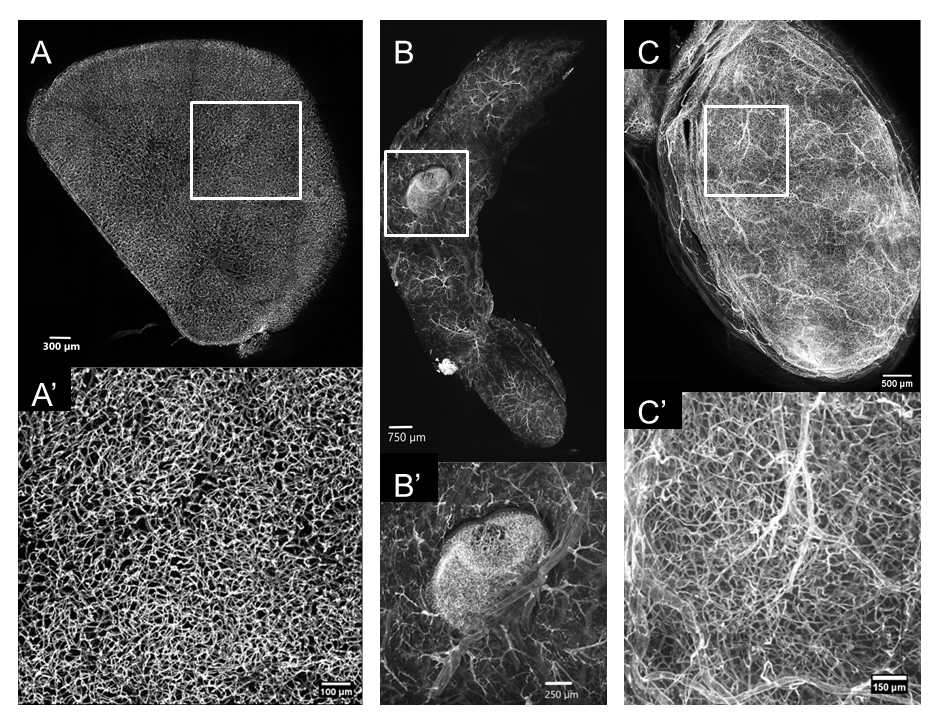
Vascular segmentation
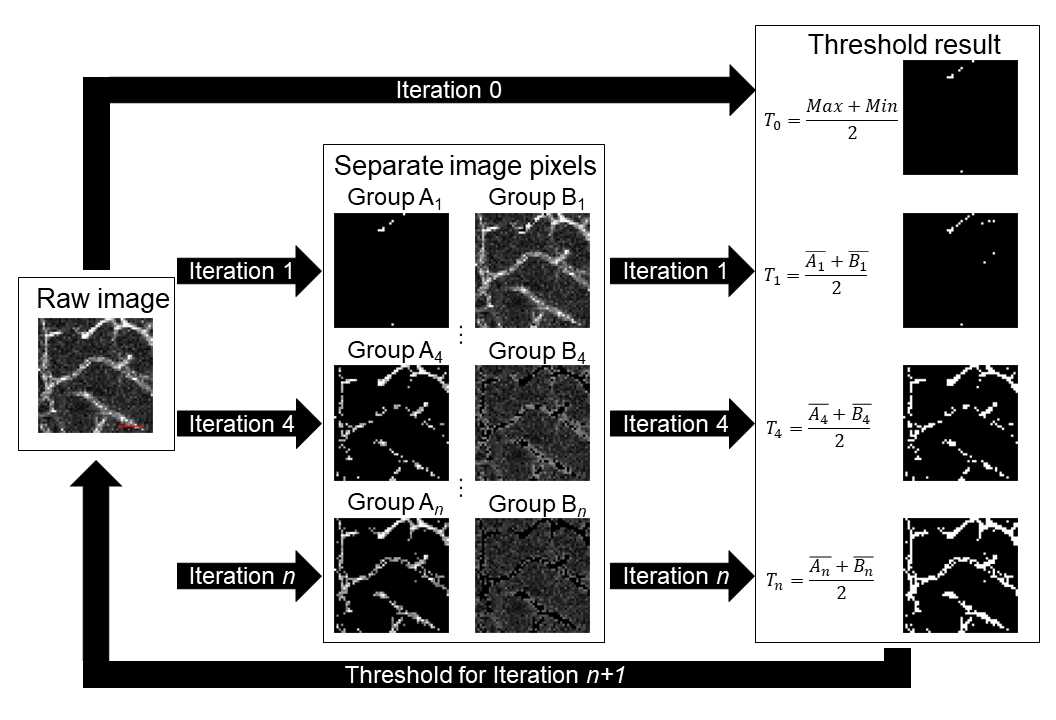
There are various segmentation methods to isolate the fluorescent vasculature from the image background. The iterative selection thresholding method was implemented to binarize the fluorescent images. This algorithm was selected due to its simplicity and objectivity and was performed using custom-written code in MATLAB.
Use a 3x3x1 median filter to remove noise within the image data. This step can be skipped if the noise in the image is sufficiently low.
The iterative selection threshold method is applied by selecting an initial threshold value, T0 0. The initial value is the average value of the maximum intensity and the minimum intensity of all voxels of an image.
All voxels within an image are separated into two groups: group An nn if a voxel has an intensity equal to or greater than the threshold value of the previous iteration, or group Bn nn if a voxel has an intensity below the threshold value of the previous iteration.
The threshold value of the subsequent iteration is calculated as the average value of the average intensity within groups An n and Bn n.
This threshold value is compared to the threshold value of the previous iteration. If the difference between the two values is less than 1, the procedure is complete, and the current threshold value is selected. If the difference between the two values is greater than or equal to 1, the procedure repeats for another iteration.
Optional morphological operations can be applied to the resulting binarized image to adjust segmentation results further. The exact parameters for these steps will vary with image acquisition parameters (particularly resolution).
neuTube tracing to quantify vessel diameters
neuTube is an open-source neuron tracing software that can be applied to tracing tubular structures, such as vasculature.
Automated tracing can be performed on binarized vasculature images in TIF file format. neuTube will output information in an SWC format where tubular structures are simplified into individual nodes with x, y, z coordinates, a radius, and node connectivity information. Each node will be a ‘parent’ to an adjacent ‘child’ node which provides the necessary information to understand how these nodes are connected in space.

