Selective Enrichment Protocol for Salmonella Isolation from Surface Water
Manan Sharma, James E Wells, Abasiofiok Mark Ibekwe, NARMS EWG, Jonathan G Frye, Autumn L. Kraft
National Program 108
Environmental Microbial and Food Safety Laboratory
Salmonella
isolation
surface water
Selective Enrichment
Abstract
This protocol describes the selective enrichment of Salmonella from surface water samples AFTER non-selective enrichment from several different recovery methods (Modified Standard Method 9260.B2, Vertical Modified Moore Swab, or Dead-end Ultra-filtration (DEUF).
Attachments
Steps
Selective Enrichment – Day 1
Dispense 9mL TT (Tetrathionate) broth into sterile test tubes.
Dispense 9mL GN (Gram-negative Hajna broth) into sterile test tubes.
Hand massage UPB(Universal Pre-enrichment Broth)-enriched modified Moore swab (MMS), BPW (Buffered Peptone Water)-enriched filter cakes (47 mm glass filters), or shake BPW-enriched bulk water or 2X BPW-enriched backflush (from DEUF filtration) to homogenize before transfers to Salmonella -selective media.
Aseptically transfer 1mL of either UPB or BPW enrichments into selective broths and then incubate at specified temperature listed in Table 1.
| A | B | C | D |
|---|---|---|---|
| Broth | Volume (mL) | Enriched UPB/BPW to add (mL) | Incubation Time and Temperature |
| TT Broth | 9 | 1 | 48 h @ 37°C |
| GN Broth | 9 | 1 | 24 h @ 37°C |
Table 1. Selective broth volumes and incubation parameters for Salmonella isolation.
Day 2 – Selective enrichment transfers
Transfer 100µL of 24 h GN broth enrichment into 9.9mL RV (Rappaport-Vassiliadis) broth.
Incubate the GN/RV enrichment at 37°C for 24h 0m 0s.
Day 3 – Selective enrichment transfers continued
Transfer 100µL of 48 h TT broth enrichment into 9.9mL RV broth.
Incubate the TT/RV enrichment at 37°C for 24h 0m 0s.
For the GN/RV enrichment, vortex tubes and use a sterile 10 µL loop to streak for isolation onto both XLT4 (Xylose Lysine Tergitol 4) (prepared with Sodium tetradecyl sulfate, XLT4 supplement) and BGS (Brilliant Green Sulfa) agar plates.
Incubate XLT4 and BGS plates at 37°C for 24h 0m 0s.
Day 4 – Selective enrichment transfers continued
For the TT/RV enrichment, vortex tubes and use a sterile 10 µL loop to streak for isolation onto both XLT4 and BGS plates
Incubate XLT4 and BGS plates at 37°C for 24h 0m 0s.
Inspect XLT4/BGS plates from Day 3 (GN/TT) for presumptive Salmonella colonies.
From the XLT4 plates, select presumptive environmental Salmonella colonies (black raised center, clear halo) and streak onto fresh XLT4 plates. Salmonella colonies on BGS appear opaque and surrounded by red color on all edges.

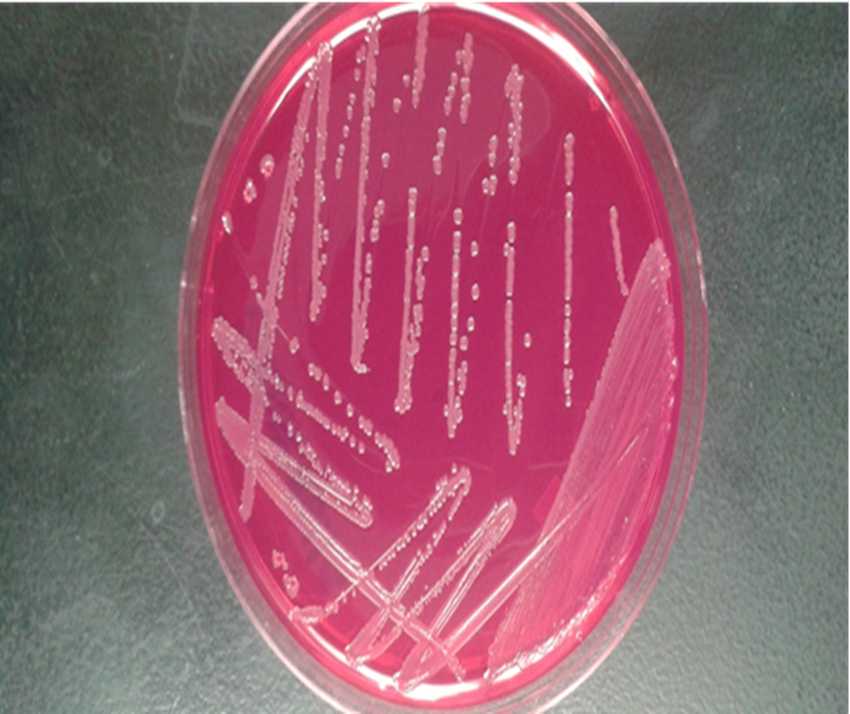
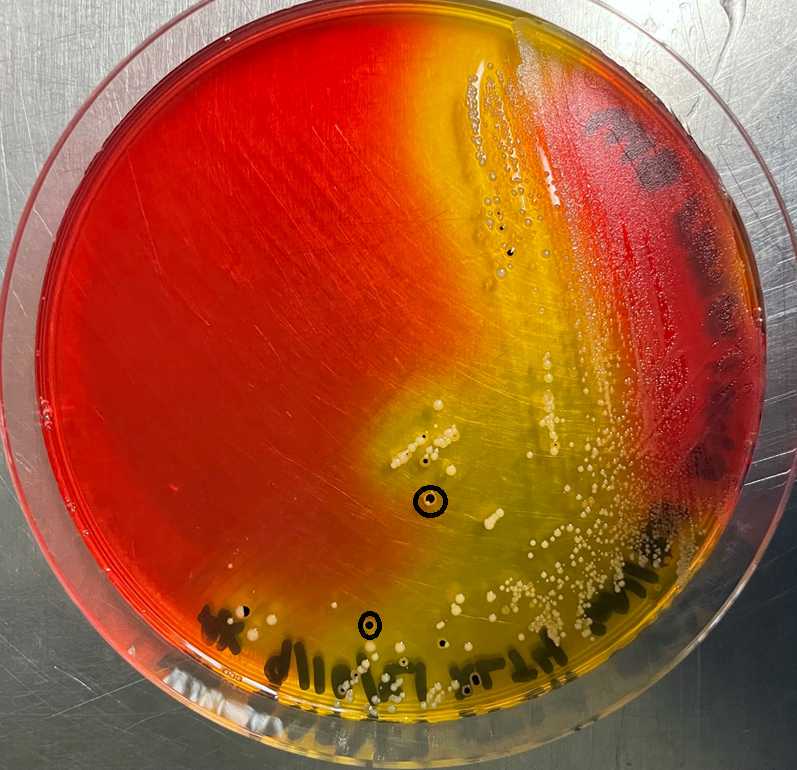
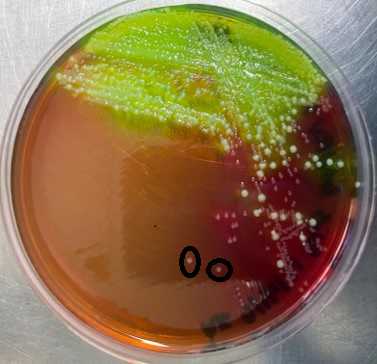
Day 5 – Colony confirmation
Inspect BGS and XLT4 plates from TT/RV enrichment and identify presumptive Salmonella colonies (see above).
Colonies from both Day 4 and 5 can be subjected to further confirmation using triple sugar iron (TSI) agar, lysine iron agar (LIA), and/or PCR (see following sections).
If non-fluorescent presumptive Salmonella isolates are observed, these isolates can be isolated and preserved (frozen stocks) for further characterization.

