SARS-CoV-2 Receptor Binding Domain Deoxy Fragment Sequencing
Noor Saber Jawad, Nuha Joseph Kandala
Abstract
Since first reporting of SARS-CoV-2 case in China, there were urgent need to report and monitor emerging variants and keep track circulating mutation, in this methodology, we used simple, cost-effective method to monitor the circulating mutations which could be used to predict the SARS-CoV-2 variants based on accumulating mutations in the receptor binding domain.
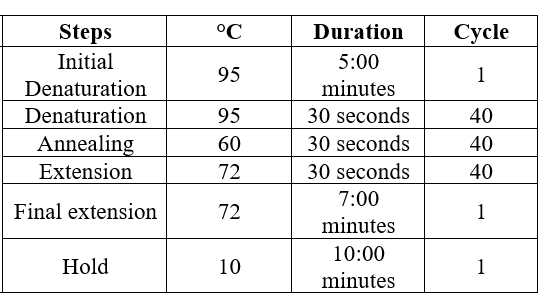
Steps
RNA Extraction
The RNA of SARS-CoV-2 extracted using Automated Extraction System ExiPrepTM 96 Lite from Bioneer (South Korea) using the Exiprep 96 Viral DNA/RNA kit using the standard recommended procedure by the manufacturer.
cDNA synthesis
The extracted RNA used as a template to construct complementary DNA, cDNA synthesis considered using the GoScriptTM Reverse Transcription Mix, Random Primers, Promega (USA) using manufacturer recommended procedure. this will ensure synthesis of the first DNA strand only.
Concentration of synthesized DNA measured by Quantus Fluorometer (Promega, USA).
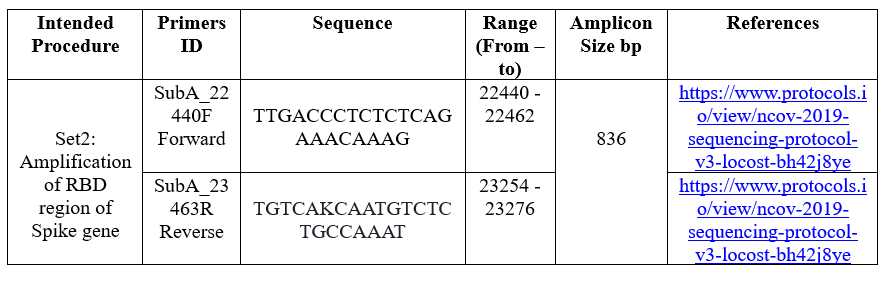
Amplicon Amplifications
RBD specific primers, amplification regions, and other details listed below:
Amplicon Sequencing
Amplicon Sequencing referred to Macrogen, South Korea to be sequenced using the ABI3730XL sequencing Machine.