SAMOSA-Tag - combining in-situ adenine methyltransferase footprinting with transposase-mediated single-molecule sequencing
Scott Nanda, Ke Wu, Siva Kasinathan, vijay.ramani
Abstract
Here we describe a protocol for SAMOSA-Tag: Tag mentation-assisted S ingle-molecule A denine M ethylated O ligonucleosome S equencing A ssay - a transposase-mediated strategy that combines our method SMRT-Tag (https://www.protocols.io/view/FB06B8000CA311ED84710A58A9FEAC02) for producing low-input PacBio sequencing libraries with in situ footprinting to enable multimodal profiling of single chromatin fibers. SAMOSA-Tag facilitates the joint analysis of genetic and epigenetic variations (i.e. somatic variation, nucleosome repeat length, CTCF occupancy, CpG methylation).
In brief, nuclei are methylated in situ using the non-specific EcoGII m6dAase, tagmented using hairpin-loaded Tn5-adaptors, gap-repaired following DNA purification, and then sequenced on the PacBio Sequel II platform. All steps from live cells to sequencing-ready libraries can be performed in one to two days. We have demonstrated the utility of SAMOSA-Tag by profiling chromatin accessibility, CpG methylation, and CTCF occupancy on low cell numbers in mammalian cell lines.

Steps
Reagent Setup
Nuclear Lysis Buffer (prepared on ice):
| A | B | C | D |
|---|---|---|---|
| Reagent (final conc.) | Stock conc. | Volume | Note |
| 20 mM HEPES | 1M, pH 7.5 | 200µL | |
| 10 mM KCl | 2M | 50µL | |
| 1 mM MgCl2 | 1M | 10µL | |
| 0.1% Triton X-100 | 10% | 100µL | |
| 20% glycerol | 100% | 2mL | |
| 1x Protease Inhibitor | 25x | 400µL | Add 25x Roche protease inhibitor immediately before use |
| dH2O | - | up to 10mL | |
| Total Volume | 10mL | ||
Buffer M (prepared on ice):
| A | B | C |
|---|---|---|
| Reagent (final conc.) | Stock conc. | Volume |
| 15mM Tris-HCl, pH 8.0 | 1M | 150µL |
| 15mM NaCl | 5M | 30µL |
| 60mM KCl | 2M | 300µL |
| 0.5mM spermidine | 1M | 5µL |
| dH2O | - | up to 10mL |
| Total Volume | 10mL | |
Nuclei Storage Buffer (prepared on ice):
| A | B | C |
|---|---|---|
| Reagent (final conc.) | Stock conc. | Volume |
| 20mM HEPES pH 7.5 | 1M | 200µL |
| 150mM NaCl | 5M | 300µL |
| 0.25mM spermidine | 1M | 2.5µL |
| 0.1X Protease Inhibitor | 25X | 40µL |
| dH2O | - | up to 10mL |
| Total Volume | 10mL | |
5x TD Premix (prepared at RT):
| A | B | C | D |
|---|---|---|---|
| Reagent (final conc.) | Stock conc. | Volume | Note |
| 10mM Tris-HCl, pH 7.5 | 1M | 10µL | |
| 5mM MgCl2 | 1M | 5µL | |
| 10% DMF | 100% | - | Add directly into Omni-ATAC Reagent Mix at step 20 |
| dH2O | - | 185µL | |
| Total Volume | 200µL | ||
SMRT-Tn5 adaptor assembly:
- we recommend following the "Annealing SMRT-Tag adaptors" and "Assembling SMRT-Tn5 transposomes (Tn5 loaded with SMRT-Tag adaptors)" procedures in SMRT-Tag protocol: https://www.protocols.io/view/FB06B8000CA311ED84710A58A9FEAC02
Nuclei isolation from fresh cells
Harvest fresh cell culture(s) in a conical centrifuge tube at room temperature and count cells. This protocol can accommodate up to a total of 2 million mammalian cells (e.g. human K562, murine embryonic stem cells) that can be divided into 10-30k nuclei per sample for up to 16 samples to be sequenced.
Centrifuge a 1 to 2 million cell suspension at 300xg at 4ºC for 5 min, and aspirate the supernatant.
[NOTE] For robust cell lines including cancer cell lines, 10 minutes of centrifugation can increase recovery.
Resuspend the cell pellet in 1mL ice-cold Nuclear Lysis Buffer. Pipet up and down gently 10x using wide-bore tips.
(Use approximately 1mL Nuclear Lysis Buffer for up to 10 million cells.)
Incubate on ice for exactly 5 min.
Centrifuge at 600xg at 4ºC for 5 min, and aspirate the supernatant.
[NOTE] For robust cell lines including cancer cell lines, 10 minutes of centrifugation can increase recovery.
Wash nuclei with 1mL Buffer M.
[cell count 0] Determine # of nuclei from a 10 µL aliquot (1:1 dilution) via hemocytomer / cell counter (10 µL nuclei suspension + 10 µL trypan blue)
Centrifuge again at 600xg at 4ºC for 5 min, and aspirate the supernatant.
[NOTE] For robust cell lines including cancer cell lines, 10 minutes of centrifugation can increase recovery.
Proceed immediately to nuclei freezing or EcoGII methylation.
(Optional) Nuclei freezing
Isolated nuclei (from step 8) may be slowly frozen by resuspending in nuclei freezing media containing 9 volumes of Nuclei Storage Buffer and 1 volume of DMSO, and further divided into 1 mL per cryogenic vial (~1-2 million nuclei), then stored at -80°C. We have found a cooling rate close to -1°C/min is sufficient for preserving nuclei integrity.
(Optional) Thawing previously frozen nuclei
If frozen nuclei are used as input, thaw ~1-2 million nuclei in a water bath at 37ºC for 1-2 min until most of the solution is a slurry. Expect 30-50% recovery after each wash step. This protocol is designed for up to 1-2 million frozen nuclei that can be further divided into 10-30k nuclei per sample for up to 16 samples to be sequenced but can be adapted for lower numbers of nuclei per replicate, or fewer replicates, as needed.
[CRITICAL] Nuclei are thawed in 10% DMSO and need to be washed rapidly in order to prevent lysis.
Use a P1000 with wide-bore pipette tips to gently transfer nuclei solution to a 1.5mL LoBind tube.
Centrifuge at 600xg at 4ºC for 5 min, and aspirate the supernatant. Leave a tiny volume around the pellet is fine.
[NOTE] For robust cell lines including cancer cell lines, 10 minutes of centrifugation can increase recovery.
Resuspend pelleted nuclei in 1mL Buffer M by gentle pipetting using a P1000 with wide-bore tips.
[cell count 1] Determine # of nuclei from a 10 µL aliquot (1:1 dilution) via hemocytomer / cell counter (10 µL nuclei suspension + 10 µL trypan blue)
Centrifuge again at 600xg at 4ºC for 5 min, and aspirate the supernatant.
[NOTE] For robust cell lines including cancer cell lines, 10 minutes of centrifugation can increase recovery.
Proceed immediately to EcoGII methylation.
in-situ SAMOSA (or EcoGII Methylation)
Resuspend the nuclei (either freshly isolated nuclei from step 8 or previoiusly-frozen nuclei from step 15) in 400 µL buffer containing Buffer M + 1mM SAM (e.g. 12.5 µL of 32mM SAM + 387.5 µL Buffer M).
[cell count 2] Determine # of nuclei from a 10 µL aliquot (1:1 dilution) via hemocytomer / cell counter (10 µL nuclei suspension + 10 µL trypan blue)
Split solution into 2 x 200 µL reactions, one for methylated (+M) and the other for unmethylated (-M) nuclei.
[NOTE] If processing a sample where unmethylated data has been previously obtained, or samples are precious, the unmethylated condition can be skipped.
[PAUSE] Determine if you have enough nuclei for downstream processing, assuming 50% loss after EcoGII methylation. (i.e 8 samples x 2 methylation states x 30k per rxn = ~ 500K nuclei required)
Add 10 µL of EcoGII (25U/µL) to +M rxn. For -M rxn, do not add EcoGII. Mix well by slowly pipetting with wide-bore tips or gently flicking to mix.
Incubate both reactions at 37ºC for 30 min in a thermomixer with 300rpm shaking every 2 min. At the 15 min time point, supplement with 1 µL of 32mM SAM stock in both reactions.
Terminate the reactions by placing them back on ice.
Tagmentation of SAMOSA-treated nuclei
Prepare Omni-ATAC Reagent Mix on ice. Prepare a little extra (5 samples worth) for sample dilution.
| A | B | C |
|---|---|---|
| Reagent (final conc.) | Stock conc. | Volume (per sample) |
| 1x TD premix | 5x | 5µL |
| 0.33x PBS | 1x | 16.5µL |
| 10% DMF | 100% | 5µL |
| 0.01% digitonin | 1% | 0.5µL |
| 0.1% Tween-20 | 10% | 0.5µL |
| dH2O | - | up to 50µL |
| Total Volume | 50µL | |
Each tagmented sample of 10-30k nuclei requires a uniquely barcoded SMRT-Tag adaptor. This protocol is designed for up to 16 samples given at least a total of 500k nuclei recovered after EcoGII methylation.
Centrifuge 2 x 200 µL reactions after EcoGII methylation at 600xg at RT for 10 min, and carefully pipette to remove the supernatant. Leave a tiny volume around the pellet is fine.
[CRITICAL] Nuclei may be smeared out across the LoBind tube wall instead of nicely pelleted at the bottom.
Resuspend nuclei gently in 250 µL Omni-ATAC Reagent Mix per reaction.
Using a P1000 with a wide-bore tip, set the pipette to 300 µL and aspirate up the entire volume per reaction. Insert the tip into a FlowMi cell strainer, and then slowly push the reaction through the strainer into a new LoBind tube. There will be bubbles ejecting upward, so make sure filtered tips are used.
[cell count 3] Determine # of nuclei post filtration from a 10 µL aliquot (1:1 dilution) via hemocytomer / cell counter (10 µL nuclei suspension + 10 µL trypan blue) for both +M and -M reactions
[CRITICAL] QC check - if nuclei still look clumped together, make a note - your count is likely NOT accurate, and may require adjusting the Tn5 amount or nuclei input per sample. Nuclei clumping tends to underestimate the actual count number.
Split each of the +M and -M reactions into multiple tubes as needed such that ~ 10K - 30K nuclei are in each tube.
[CRITICAL] The majority of samples to be sequenced should be +M which are used to profile chromatin accessibility.
To generate an average of 3kb libraries, add 2 µL of loaded SMRT-Tn5 adaptor stock (~ 9.4µM monomer) per sample and fill the total volume up to 50 µL with Omni-ATAC reagent mix.
[OPTIONAL] QC check - Prepare 1 untagged negative control for each of +M and -M reactions to compare against tagged samples. A total of two negative controls: +M -tag and -M -tag. Replace SMRT-Tn5 adaptor volume with Omni-ATAC reagent mix.
Incubate all tagmentation reactions at 55ºC for 1 hr and hold at 4ºC.
Tagmentation termination
Treat the tagmented nuclei with 10 µL of RNase A (10ug/µL) and incubate at 37ºC for 15 min on a thermomixer with 300rpm mixing every 2 min.
[CRITICAL] RNAse pre-treatment is critical to ensure that only DNA is extracted from the reaction.
Prepare the Termination Lysis Reagent Mix about 10 min before the tagmentation is complete.
| A | B | C |
|---|---|---|
| Reagent (final conc.) | Stock conc. | Volume (per sample) |
| Proteinase K (6.66mg/mL) | 20mg/mL | 2.5µL |
| 3.33% SDS | 10% | 2.5µL |
| 167mM EDTA | 0.5M | 2.5µL |
| Total Volume | 7.5µL |
[CRITICAL] Equilibriate Proteinase K at RT before adding to the mixture. Do NOT keep it on ice, or the SDS mixture will precipitate with EDTA. Vortex EDTA stock solution prior to use. If the mixture appears cloudy, warm it up at RT until it appears homogenous again.
After RNase treatment, add 7.5 µL of termination lysis reagent mix to each sample and mix well.
Incubate at 60ºC on a thermomixer with 1000rpm continuous shaking for at least 1 hr, up to 2 hr for improved lysis.
gDNA purification with SPRI cleanup
Remove samples from thermomixer and incubate on ice for 1 min to cool down samples to RT.
Add 2x volume of resuspended, room-temperature SPRI beads. (i.e. For a 50 µL tagmentation reaction, sample volume should be 50 µL tagmentation reaction + 10 µL RNase A + 7.5 µL termination lysis MM ~ 67.5 µL, so 2X ~ 135 µL). Mix well using a P200 with wide-bore tips.
Incubate the bead-mixed samples in a thermomixer at 23ºC for 30 min with interval mixing @350 rpm (1 min on, 3 min off) to keep the beads resuspended.
Spin down quickly. Place on magnet and allow to clear before carefully withdrawing supernatant.
Wash beads twice with 80% ethanol for 30 sec. Add ethanol to the wall opposite of beads. Withdraw supernatant and after a quick spin, remove the remaining liquid with a P20 pipette. Beads take ~ 30 s to 1 min to dry. Do not over-dry the beads - dried beads appear to be fragmented with cracks in light brown.
Remove from the magnet stand, and gently resuspend the bead pellet in 20 µL 1x EB (or 10 mM Tris-HCl, pH 8.5) to elute the extracted DNA. Once beads are resuspended, mix well.
Incubate the samples in a Thermomixer at 37ºC for 15 min with interval mixing @350 rpm (1 min on, 3 min off) to promote increased DNA elution.
Spin samples down quickly. Place on magnet and allow samples to clear before carefully transferring the supernatant to a new LoBind tube.
[Pause] Purified gDNA samples can be stored at 4ºC for up to two weeks.
Determine concentrations of purified DNA using Qubit 1x High Sensitivity DNA Assay. Expect at least 160ng of DNA recovered from 10-30k nuclei.
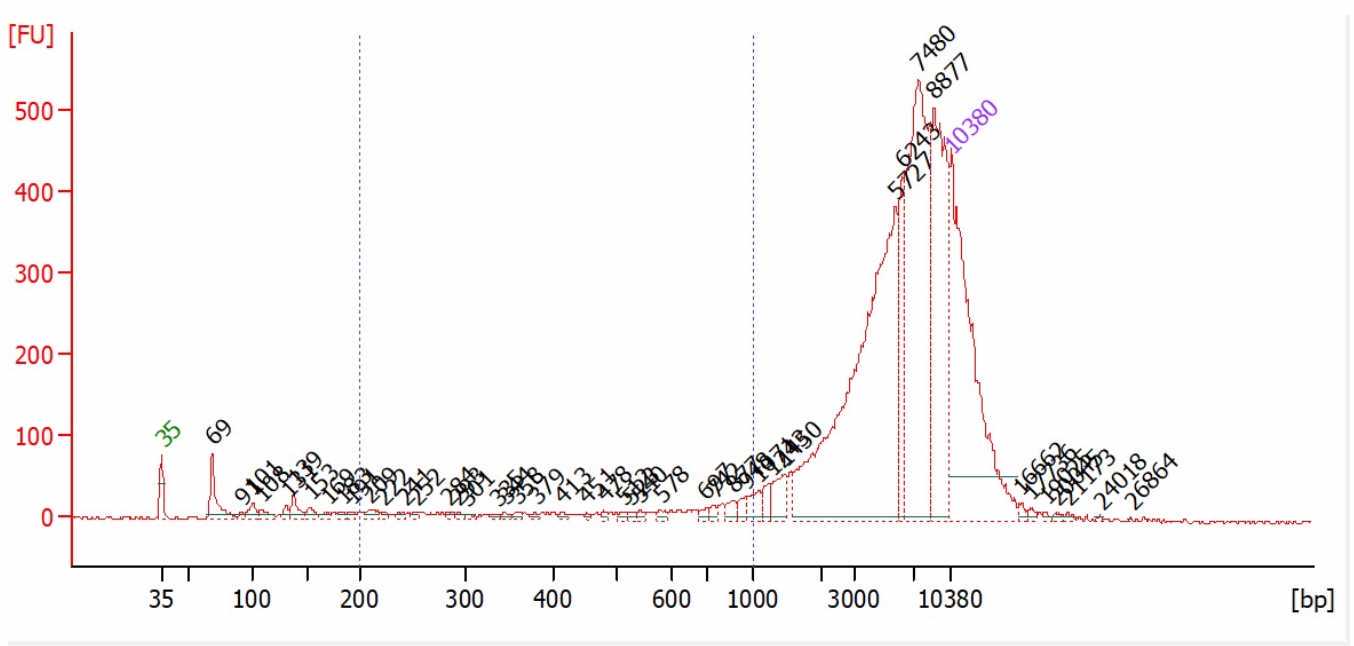
[OPTIONAL] Determine the fragment size distribution using Agilent 2100 Bioanalyzer High Sensitivity DNA Assay.
[OPTIONAL] If BioAnalyzer trace shows a trailing tail <1kb (e.g. reference fig. 1), we recommend doing an additional 0.6x Ampure PB cleanup to enrich for fragments > 500bp.
[Optional] QC check - Visualize the SMRT-Tn5 transposome efficiency by running 2 untagged controls against tagged samples on a 0.5% agarose gel coupled with the NEB 1kb-Extend ladder. Expect to see the majority of bands clustered towards high molecular weight >48.5kb for untagged samples, whereas a long smear should be observed for tagged samples. Untagged samples will not be sequenced as they do not contain SMRT-Tag adaptors.
Gap Repair
Prepare the Gap-Repair Reagent Mix.
| A | B | C |
|---|---|---|
| Reagent (final conc.) | Stock conc. | Volume (per sample) |
| tagmented sample (up to 160ng) | - | 12µL* |
| dNTP mix (0.8mM) | 8mM | 2µL |
| 1x Taq DNA Ligase Rxn Buffer | 10x | 2µL |
| NEB Phusion (0.1U) | 2U/µL | 1µL |
| NEB Taq DNA Ligase (4U) | 40U/µL | 2µL |
| dH20 | - | 1µL |
| Total Volume | 20µL | |
*Normalize each tagmented sample to 160ng in 12µL with 1x EB.
Incubate the gap-repair reactions at 37ºC for 1hr and hold at 4ºC.
Add 2x volume of resuspended, room-temperature SPRI beads to clean up the reactions. Mix well using a P200 with wide-bore tips.
Incubate the bead-mixed samples in a thermomixer at 23ºC for 15 min with interval mixing @350 rpm (1 min on, 3 min off) to keep the beads resuspended.
Spin down quickly. Place on magnet and allow to clear before carefully withdrawing supernatant.
Wash beads twice with 80% ethanol for 30 sec. Add ethanol to the wall opposite of beads. Withdraw supernatant and after a quick spin, remove the remaining liquid with a P20 pipette. Beads take ~ 30 s to 1 min to dry. Do not over-dry the beads - dried beads appear to be fragmented with cracks in light brown.
Remove from the magnet stand, and gently resuspend the bead pellet in 12 µL 1x EB (or 10 mM Tris-HCl, pH 8.5) to elute the extracted DNA. Once beads are resuspended, mix well.
Incubate the samples in a Thermomixer at 37ºC for 10 min with interval mixing @350 rpm (1 min on, 3 min off) to promote increased DNA elution.
Spin samples down quickly. Place on magnet and allow samples to clear before carefully transferring the supernatant to a new LoBind tube.
[Optional] QC check - take 1 µL aliquot and determine concentrations of gap-repaired samples using Qubit 1x High Sensitivity DNA Assay. Expect a 70% or above recovery from tagmented sample input in step 44.
Exonuclease Digestion
Prepare the Exo-Digest Reagent Mix.
| A | B | C |
|---|---|---|
| Reagent (final conc.) | Stock conc. | Volume (per sample) |
| gap-repaired sample | - | 12µL* |
| NEBuffer 2 (1x) | 10x | 1.5µL |
| Exonuclease III (100U) | 100U/µL | 1µL |
| dH20 | - | 0.5µL |
| Total Volume | 15µL | |
*Diluted up to 12µL with 1x EB for volume <12µL.
Incubate the exo-digested reactions at 37ºC for 1hr and hold at 4ºC.
Add 2x volume of resuspended, room-temperature SPRI beads to clean up the reactions. Mix well using a P200 with wide-bore tips.
Incubate the bead-mixed samples in a thermomixer at 23ºC for 15 min with interval mixing @350 rpm (1 min on, 3 min off) to keep the beads resuspended.
Spin down quickly. Place on magnet and allow to clear before carefully withdrawing supernatant.
Wash beads twice with 80% ethanol for 30 sec. Add ethanol to the wall opposite of beads. Withdraw supernatant and after a quick spin, remove the remaining liquid with a P20 pipette. Beads take ~ 30 s to 1 min to dry. Do not over-dry the beads - dried beads appear to be fragmented with cracks in light brown.
Remove from the magnet stand, and gently resuspend the bead pellet in 12 µL 1x EB (or 10 mM Tris-HCl, pH 8.5) to elute the extracted DNA. Once beads are resuspended, mix well.
Incubate the samples in a Thermomixer at 37ºC for 10 min with interval mixing @350 rpm (1 min on, 3 min off) to promote increased DNA elution.
Spin samples down quickly. Place on magnet and allow samples to clear before carefully transferring the supernatant to a new LoBind tube.
Library QC and Sequencing
Determine library concentration using Qubit 1x High Sensitivity DNA Assay.
Determine the size distribution using Agilent 2100 Bioanalyzer High Sensitivity DNA Assay.
[CRITICAL] Convert the BioA trace from its default unit (fluorescent unit) to "molarity per length" using the R package "bioanalyzeR". This step is critical for the evaluation of accurate library sizing required for the optimal loading on PacBio Sequel II.
Pool barcoded libraries equimolarly and sequence on PacBio Sequel II 8M SMRTcells. We recommend using Sequel II Binding Kit 2.1 for the best loading result, as well as a 30h movie time, 2h pre-extension, and 4h immobilization time.