Running the Titan_ClearLabs Workflow on Terra.bio
Jill V Hagey, Kevin Libuit, Technical Outreach and Assistance for States Team, Frank J Ambrosio
ONT
Nanopore
SARS-CoV-2
MinION
GridION
PromethION
MK1C
Pangolin
Genomics
Analysis
Virology
Bioinformatics
RNA
DNA
Clear Labs
Covid
Computational Biology
Sequencing
Clear Labs
Hamilton
Disclaimer
The opinions expressed here do not necessarily reflect the opinions of the Centers for Disease Control and Prevention or the institutions with which the authors are affiliated. The protocol content here is under development and is for informational purposes only and does not constitute legal, medical, clinical, or safety advice, or otherwise; content added to protocols.io is not peer reviewed and may not have undergone a formal approval of any kind. Information presented in this protocol should not substitute for independent professional judgment, advice, diagnosis, or treatment. Any action you take or refrain from taking using or relying upon the information presented here is strictly at your own risk. You agree that neither the Company nor any of the authors, contributors, administrators, or anyone else associated with protocols.io, can be held responsible for your use of the information contained in or linked to this protocol or any of our Sites/Apps and Services.
Abstract
The Titan_ClearLabs workflow is a part of the Public Health Viral Genomics Titan series for SARS-CoV-2 genomic characterization. Titan_CleanLabs was written to process Clear Labs read data for SARS-CoV-2 ARTIC V3 amplicon sequencing. Upon initiating a Titan_ClearLabs run, input read data provided for each sample will be processed to perform consensus genome assembly, infer the quality of both raw read data and the generated consensus genome, and assign lineage or clade designations as outlined in the Titan_ClearLabs data workflow below.
Additional technical documentation for the Titan_ClearLabs workflow is available at:
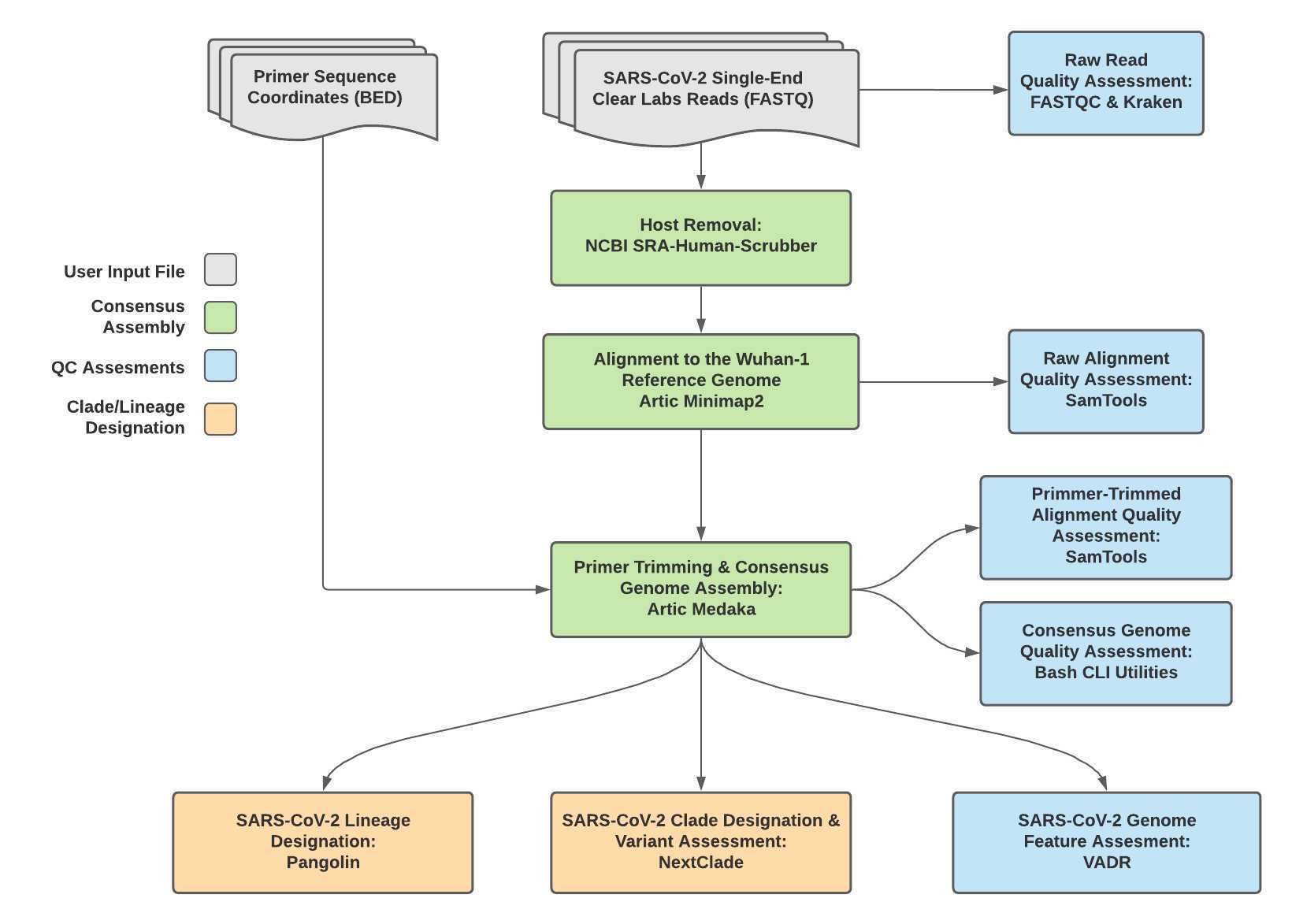
Required input data for Titan_ClearLabs:
Cear Labs FASTQ read files (single FASTQ file per sample)
Primer sequence coordinates of the PCR scheme utilized in BED file format
Video Instruction:
Theiagen Genomics: Titan Genomic Characterization
https://www.youtube.com/watch?v=zP9I1r6TNrw
Theiagen Genomics: Titan Outputs QC
https://www.youtube.com/watch?v=Amb-8M71umw
For technical assistance, please contact us at: TOAST@cdc.gov
Attachments
Steps
Running the Titan_ClearLabs Workflow
To run the Titan_ClearLabs workflow, click on the 'Workflows' panel in the newly created workspace. It should bring you to your workflow page. Click on the 'Titan_ClearLabs' tile to bring up the Titan_ClearLabs assembly workflow page (if you do not see the Titan_ClearLabs workflow in your workspace, please see our video on importing a workflow to Terra.bio: https://youtu.be/ZRpQylIDMzo).
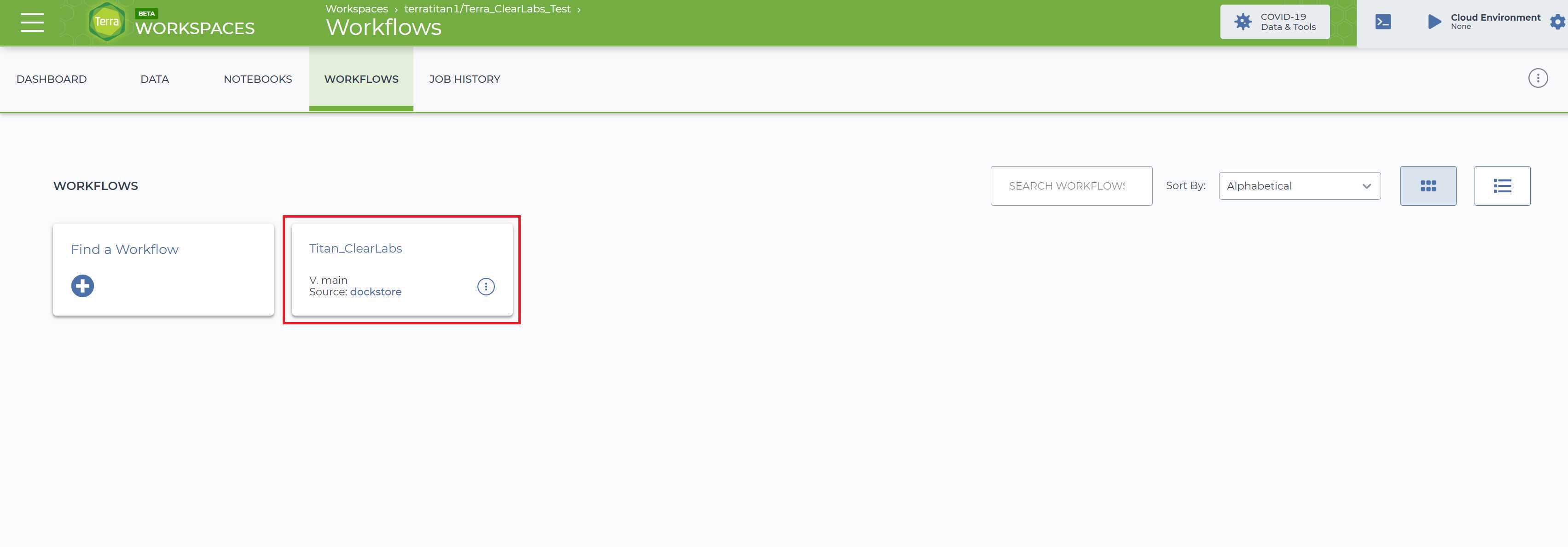
This will bring up the Titan_ClearLabs workflow page:
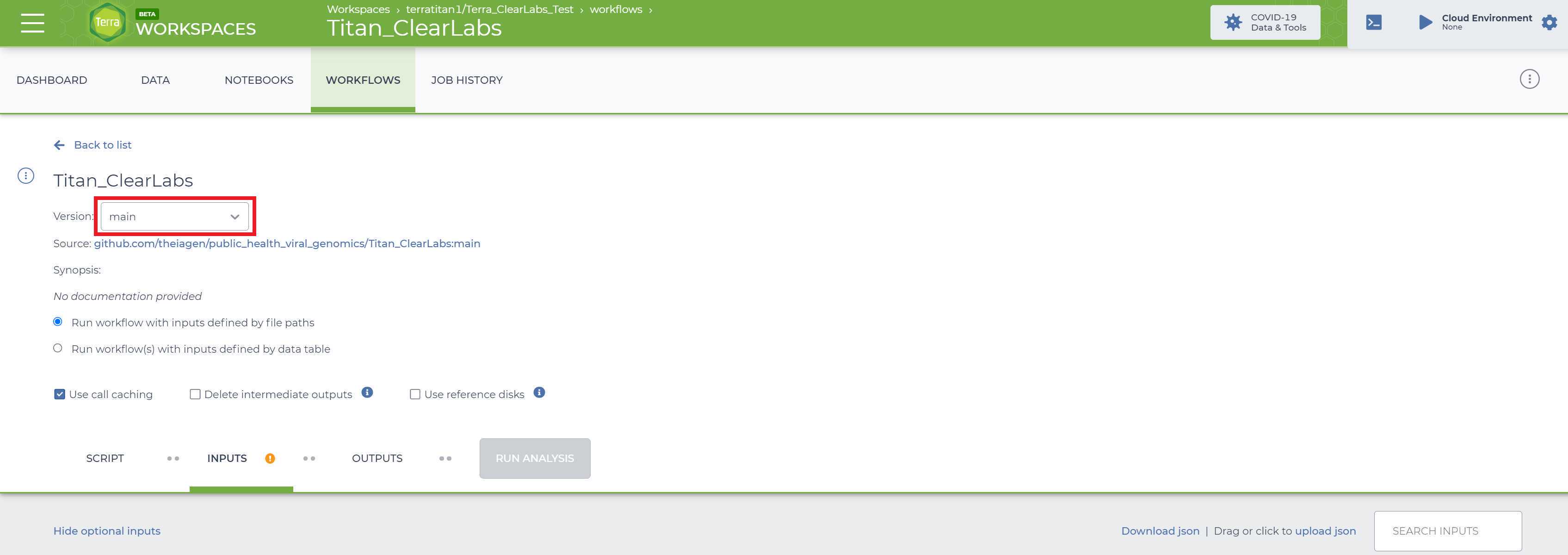
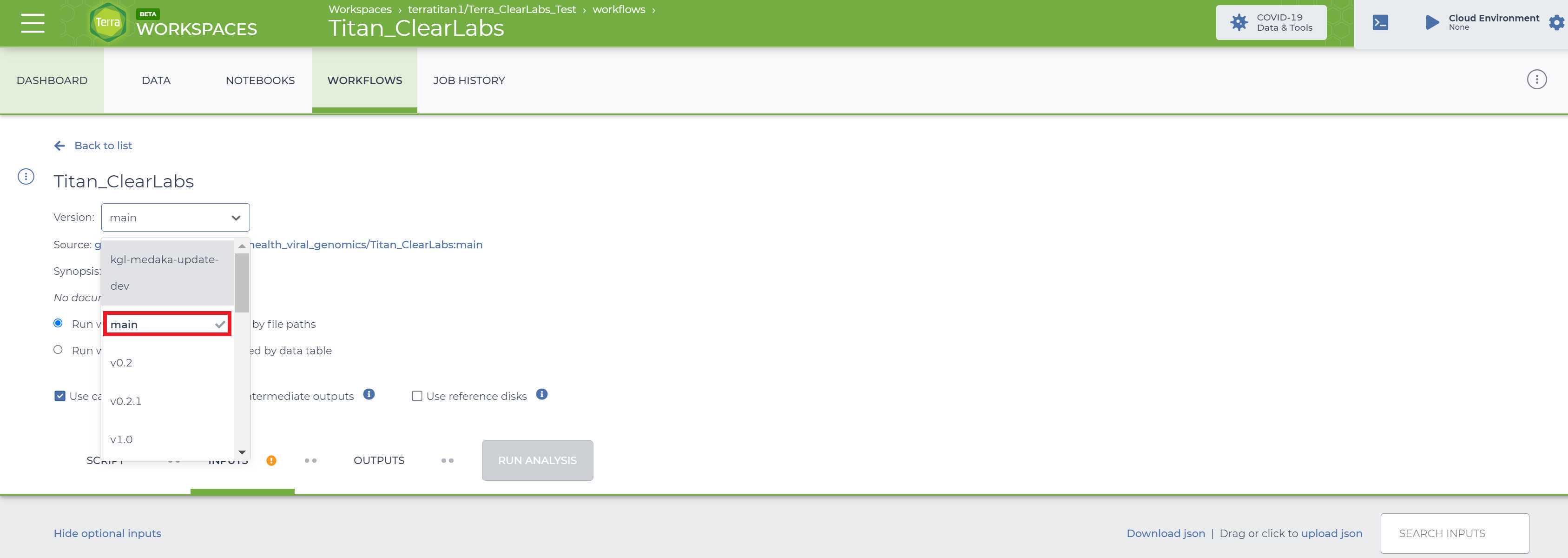
Select the version of the workflow you would like to run. Double check that you are using the latest version of the workflow. Alternately, you may specify another version, but should only pick a stable version (with numbers) NOT a 'main' or 'dev' version.
Ensure that "Run workflow(s) with inputs defined by data table" is selected and the "Use call caching" is checked and then select the root entity type for the data you wish to analyze
NOTE: Call caching allows Terra to identify and skip jobs that have been run previously; this option is by default enabled to avoid unnecessary compute costs. More information on Terra call caching, including examples of when you may want to disable this feature, is available through the Terra Support Documentation.
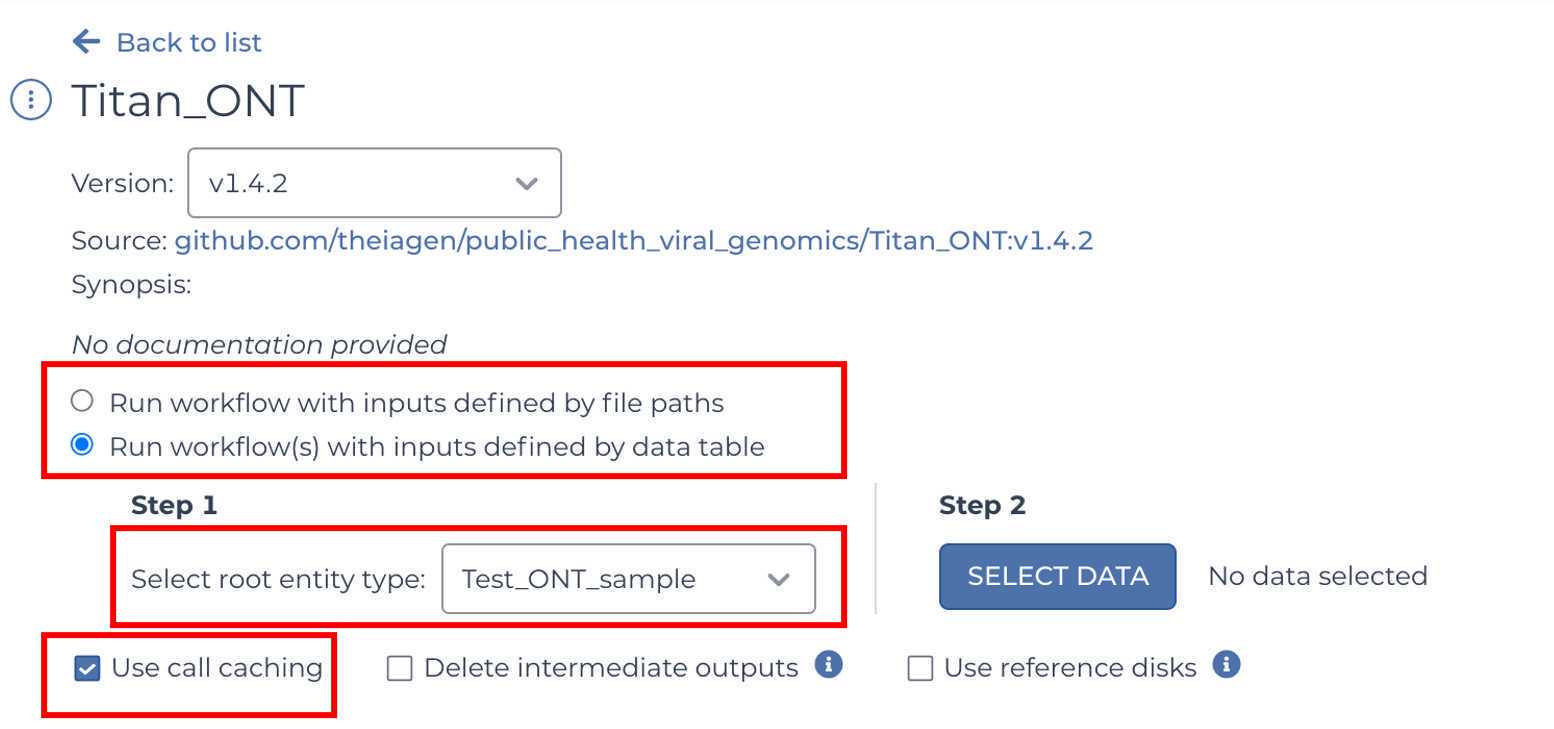
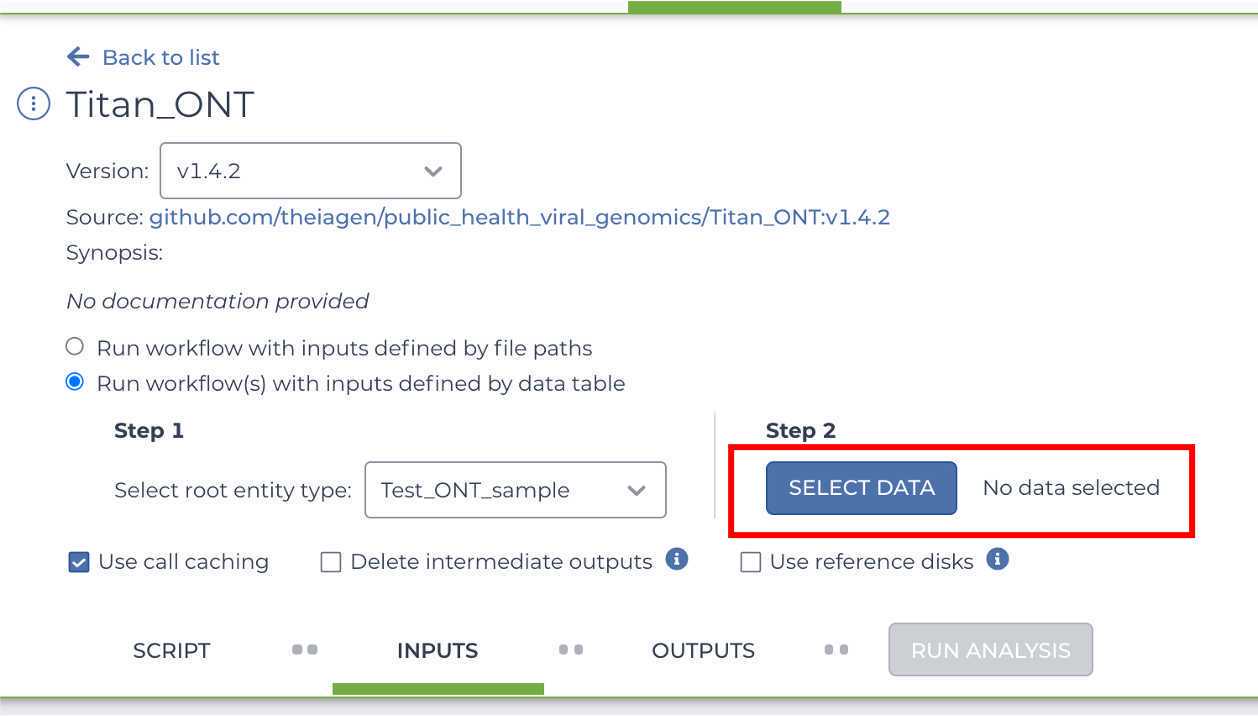
Click "SELECT DATA" and choose the samples you wish to analyze
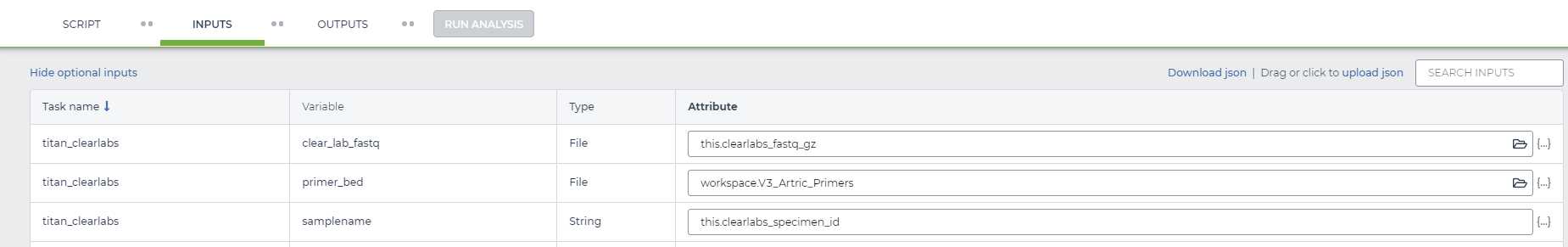
Complete the INPUTS form with the appropriate attributes (if you have not yet uploaded any Clear Labs data to Terra.bio please reach out to the Theiagen Genomics bioinformatics support team [support@terrapublichealth.zendesk.com] and we will help you to establish Terra.bio integration with the Clear Labs portal")
The top two rows represent variables that have to be provided by the user. This was the information that we populated the sample data table with in the previous step.
In our example, for the first row, the 'clear_lab_fastq' variable, we clicked on the 'Attribute' text box and wrote 'this.clearlabs_fastq_gz' to indicate that the 'clear_lab_fastq' we wish to analyze are under the 'clearlabs_fastq_gz' column of our selected datatable. In the second row we input our primer_bed file of choice, which should be saved as a workspace element in Terra (if you do not have this saved as a workspace element please see our video on uploading a workspace element: https://youtu.be/Qrbz7uRtwmQ). In the third row, the 'samplename' variable, we selected on the 'Attribute' text box and wrote 'this.clearlabs_specimen_id' to indicate the 'samplename' of each sample we are analyzing can be found in the `Test_ONT_sample_id' column of our selected datatable.
NOTE: If you named your columns something other than reads then just type "this." followed by whatever the column name is. We would advise naming your reads column "reads" for clarity.
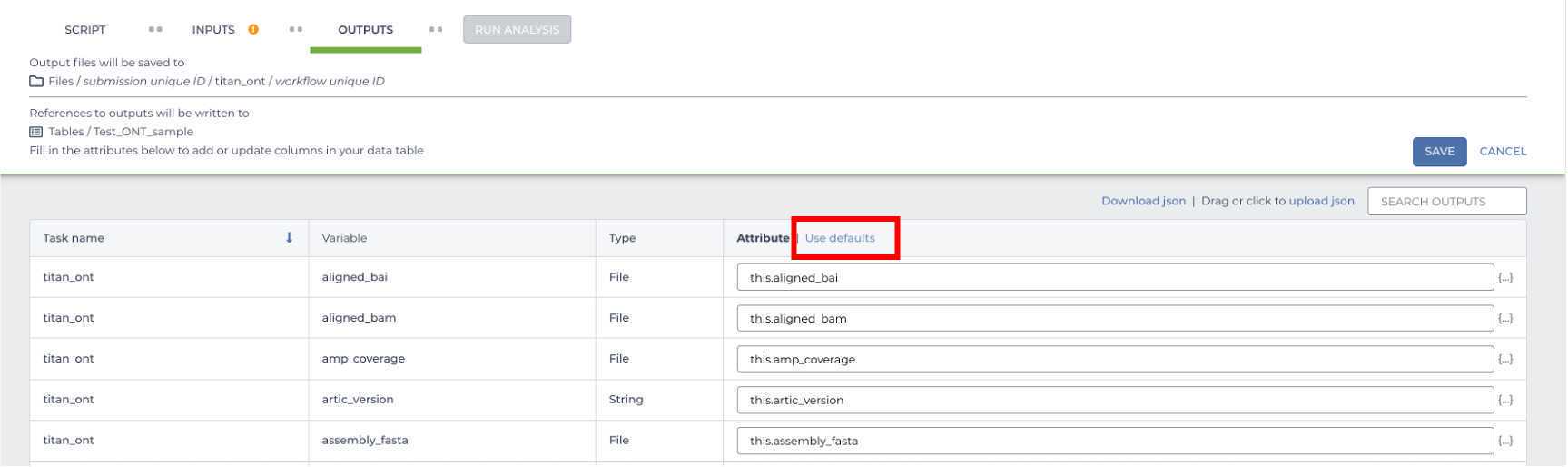
Once your input form is complete, move on to the OUTPUTS form and select "Use Defaults'. Terra will then populate the OUTPUTS form with all of the default outputs options generated by the workflow. If you forget to do this you won't have easily accessible results! Save these changes by clicking the 'Save' button.
Once your INPUTS and OUTPUTS forms are complete, click the 'Save' button on the top right-hand side of the page. The yellow caution icons should disappear and the Run Analysis option should be made available.
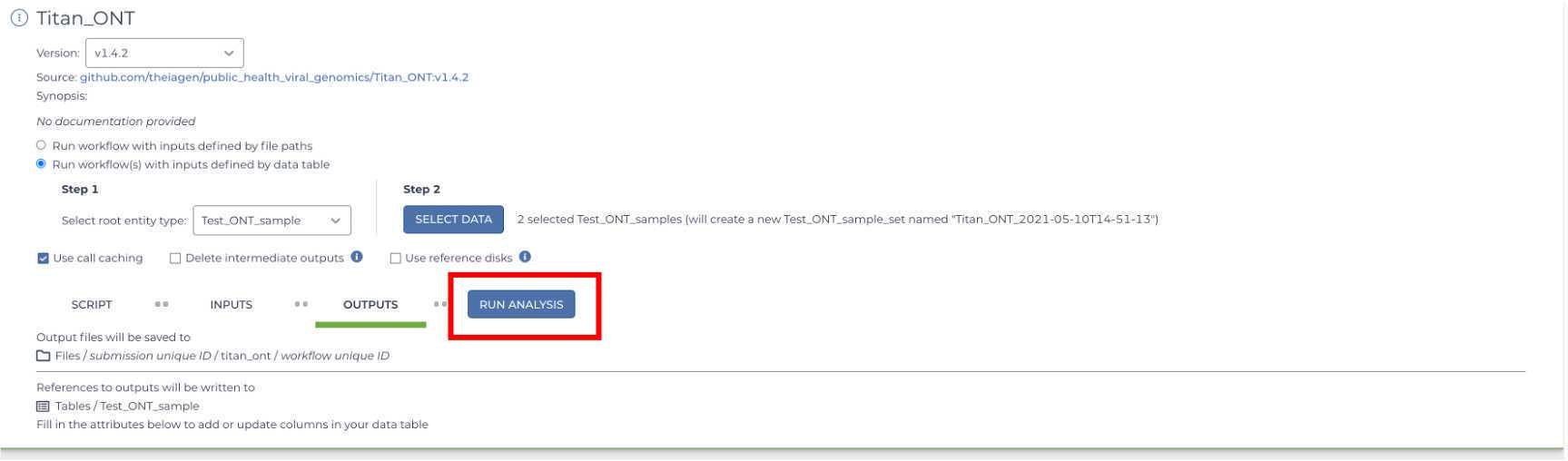
You are now ready to run the Titan_ClearLabs workflow! Click on the 'Run Analysis' button to the right of the 'Outputs' tab. A popup window should appear titled 'Confirm launch'. If the 'Run Analysis' button is greyed out, you need to save your recent changes by clicking the 'Save' button.
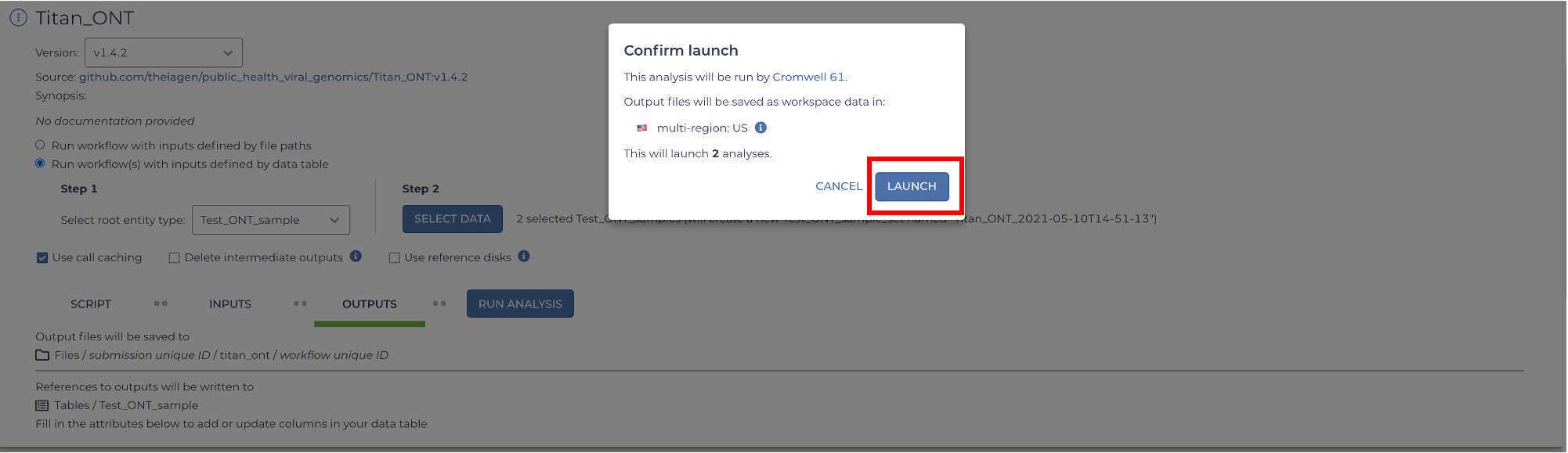
Clicking the 'Launch' button should bring you to the 'Job History' panel where each sample will be queued for the Titan_ClearLabs analysis. The status will change from queued to submitted to running.
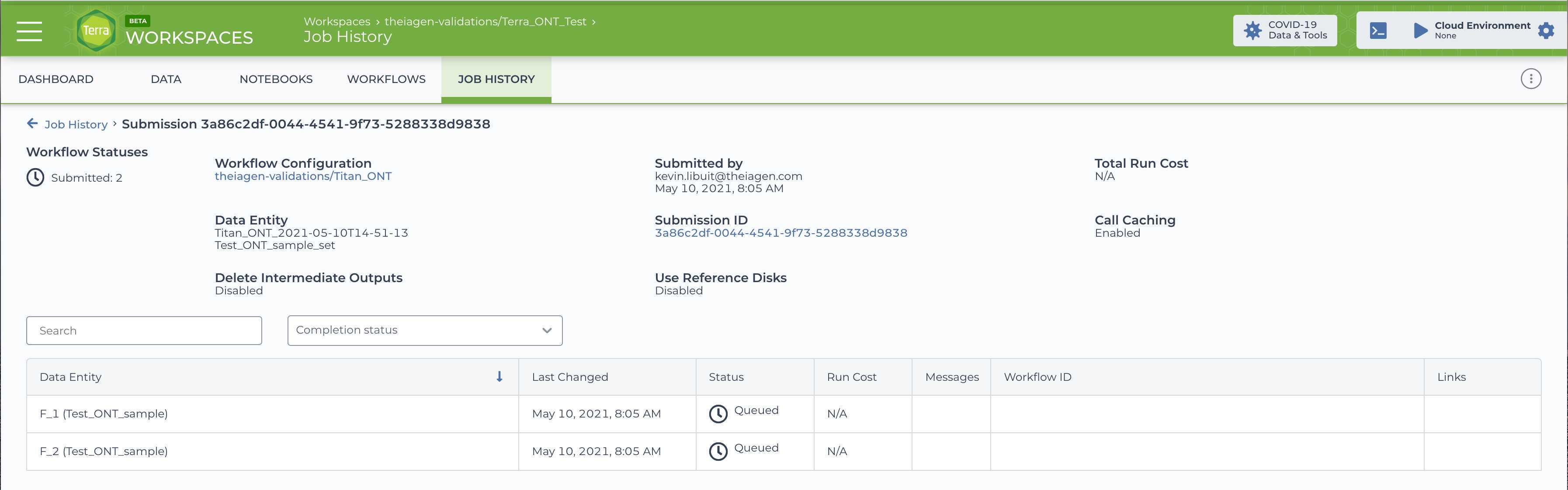
Any trouble at all please feel free to check out our video on Running the Titan_ClearLabs Workflow, or reach out to our support email: support@terrapublichealth.zendesk.com

