Roadmap to the bioinformatic study of gene and protein evolution
florian.jacques, Paulina Bolivar, Kristian Pietras
Evolution
bioinformatics
phylogenetic analysis
evolutionary studies
molecular evolution
Phylogenetic inference
Abstract
We present a compilation of nucleic acid and protein databases and bioinformatic tools for phylogenetic reconstructions and a wide range of studies on molecular evolution and population genetics. We provide a protocol for information extraction from biological databases and simple phylogenetic reconstruction.
Steps
Sequence selection and comparisons
State of the art on genes and proteins
Evolutionary analyses on molecular data, (genes, genomes or proteins), generally require retrieving information from public biological databases. Several dozens of databases store information about the state of the art on genes and proteins. In particular, many of them provide the sequences in fasta format, which is necessary for evolutionary studies. Other information and annotations, including structure, activity, biological function, tissue expression, subcellular location and polymorphism can also prove relevant.
Here is a non-exhaustive list of nucleic acid databases, and a list of the main protein databases, with their main features.
Retrieve the sequence from one of the following databases (e.g. NCBI or Uniprot) and paste the sequence in a fasta file using fasta format, using .fasta as filename extension.
Fasta format includes a headline starting with ">", and the nucleic acid or amino acid sequence.
For example:
sp|P04637|P53_HUMAN Cellular tumor antigen p53 OS=Homo sapiens OX=9606 GN=TP53 PE=1 SV=4 MEEPQSDPSVEPPLSQETFSDLWKLLPENNVLSPLPSQAMDDLMLSPDDIEQWFTEDPGP DEAPRMPEAAPPVAPAPAAPTPAAPAPAPSWPLSSSVPSQKTYQGSYGFRLGFLHSGTAK SVTCTYSPALNKMFCQLAKTCPVQLWVDSTPPPGTRVRAMAIYKQSQHMTEVVRRCPHHE RCSDSDGLAPPQHLIRVEGNLRVEYLDDRNTFRHSVVVPYEPPEVGSDCTTIHYNYMCNS SCMGGMNRRPILTIITLEDSSGNLLGRNSFEVRVCACPGRDRRTEEENLRKKGEPHHELP PGSTKRALPNNTSSSPQPKKKPLDGEYFTLQIRGRERFEMFRELNEALELKDAQAGKEPG GSRAHSSHLKSKKGQSTSRHKKLMFKTEGPDSD
| A | B | C |
|---|---|---|
| Database | Features | Link |
| BAR | Database of plant genes and proteins | http://bar.utoronto.ca/ |
| Bgee | Gene expression patterns | https://bgee.org/ |
| Ensembl | Genome browser of vertebrates, includes tools for identification of homology | https://www.ensembl.org/index.html |
| Entrez | Gene sequences and structures | https://www.ncbi.nlm.nih.gov/Web/Search/entrezfs.html |
| FigTree | Vizualization tool for phylogenetic trees | https://github.com/rambaut/figtree/releases |
| FlyBase | Genome and proteins of the model insect D. melanogaster | https://flybase.org/ |
| GenBank | Annotated DNA sequences | https://www.ncbi.nlm.nih.gov/genbank/ |
| PomBase | Genes and proteins of the model yeast S. pombe | https://www.pombase.org/ |
| TAIR | Genome and proteins of the model plant A. thaliana | https://www.arabidopsis.org/ |
| WormBase | Genome and proteins of the model nematode C. elegans | https://wormbase.org//#012-34-5 |
| Xenbase | Genome and proteins of the model amphibian X. laevis | http://www.xenbase.org/entry/ |
List of the main nucleic acid databases
| A | B | C |
|---|---|---|
| Database | Features | Link |
| Gene ontology | Unified annotation of molecular function, biological processes, and cellular components of proteins | http://geneontology.org/ |
| Interpro | Classification of proteins domains and functional sites | https://www.ebi.ac.uk/interpro/ |
| KEGG | Protein function and biological pathways | https://www.genome.jp/kegg/ |
| PDB | 3-dimensional structures of proteins | https://www.rcsb.org/ |
| Pfam | Information about protein families and domains, includes tools for identification of homology | http://pfam.xfam.org/ |
| Pharos | Centralizes literature for human proteins | https://pharos.nih.gov/ |
| Prints | Protein fingerprints classification database | http://www.bioinf.man.ac.uk/dbbrowser/PRINTS/ |
| Prosite | Protein family database | https://prosite.expasy.org/ |
| Scop | Classification of protein based on their structure | https://scop.mrc-lmb.cam.ac.uk/ |
| Superfamily | Protein structure and functions | https://supfam.org/ |
| The human protein atlas | Information on human protein and their link with diseases | https://www.proteinatlas.org/ |
| Uniprot | General information on proteins | https://www.uniprot.org/uniprot/P04637 |
List of the main protein databases
Protein classification
Using protein classification can also be relevant for evolutionary studies. Proteins are classified into different categories based on structural and functional similarity, evolutionary relationship, or both. Retrieving the classification of a protein of interest, and identifying the main protein domains, often provides valuable insight on its biodiversity and evolutionary origin. Several classification systems are published and listed in the table above.
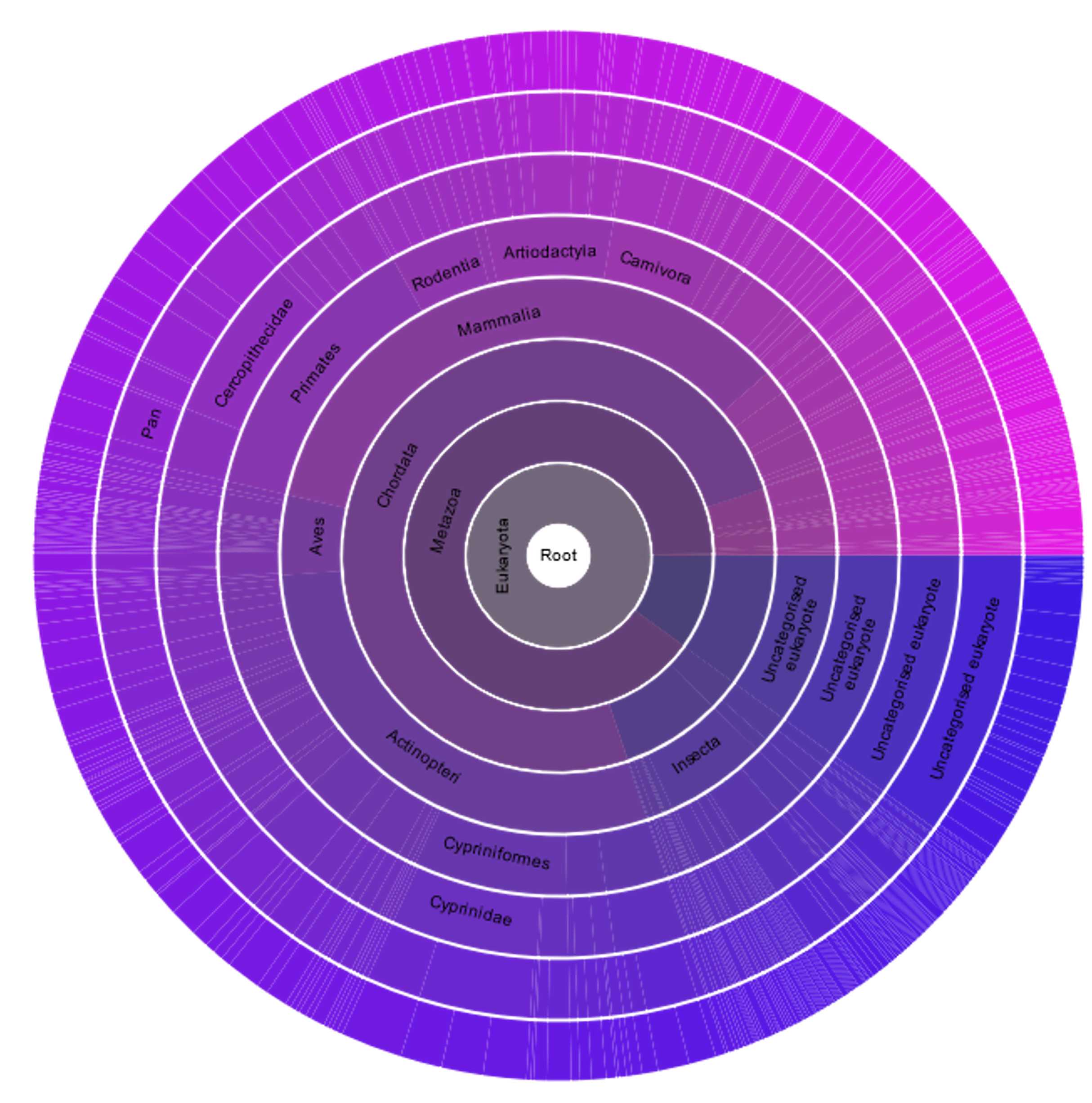
Use a classification system (e.g. Pfam or Interpro) to identify the main domains of a protein. Pfam presents also their occurrence in living organisms as a sunburst plot.
This figure displays the diversity of the domain P53 that can be retrieved from Pfam. It shows that this domain is present in virtually all animals, and some of their close relatives, such as choanoflagellates, and suggests that this domain appeared before the divergence between animals and these protists.
Identification of homologues
Studying the evolution of a family of genes or proteins requires the identification of homologues, i.e. , genes or protein with shared ancestry. Homologues include orthologues, that are present in different species, and paralogues, that are present in the same genome. Bioinformatic tools can be used to identify gene or protein homology based on sequence similarity, in the genomes of any species (see the list below).
Use the sequence of your gene of interest under fasta format to identify homologues in the genomes of other species using BLAST, for example. Paste the sequences of the homologues in a single fasta file and select the species or groups of species of interest (e.g. all animals and choanoflagellates). Download the sequences of the homologues covering the diversity of animals and paste them under fasta format in another fasta file.
Here is a list of tools that can be used to identify sequence homologues:
| A | B | C |
|---|---|---|
| Database | Features | Link |
| BLAST | Protein or DNA homology search | https://blast.ncbi.nlm.nih.gov/Blast.cgi |
| Blat | Sequence search | https://genome.ucsc.edu/cgi-bin/hgBlat |
| Ensembl | DNA homology search | https://m.ensembl.org/index.html |
| FASTA | Sequence search and alignment | https://www.ebi.ac.uk/Tools/sss/fasta/ |
| HMMER | Protein homology search | http://hmmer.org/ |
| Shaha | Sequence search and alignment | https://www.sanger.ac.uk/tool/ssaha/ |
List of bioinformatic tools for identification of gene and protein homologues
Sequence alignment
Phylogenetic analysis requires identification of homologue bases or amino acid residues of the different sequences. Homology is inferred by a sequence alignment. Sequences of different genes or proteins are put in every row one after the other to arrange every homologous base or amino acid in the same column.
Here is a list of tools for nucleic acid or amino acid sequence alignment. Use an appropriate tool (nucleic acid or amino acid) to generate an alignment of the sequences. Insertions and deletions are indicated by gaps "-" added to the sequences.
It is sometimes recommended to check the alignment and, when necessary, to improve it manually or using GBlocks. Then, export the alignment using fasta format.
| A | B | C |
|---|---|---|
| Software | Features | Link |
| BAli-Phy | Bayesian alignment and phylogenetic analysis | http://www.bali-phy.org |
| ClustalOmega | DNA or amino acid sequence alignment | https://www.ebi.ac.uk/Tools/msa/clustalo/ |
| ClustalW | DNA or amino acid sequence alignment | https://www.genome.jp/tools-bin/clustalw |
| GBlocks | DNA or amino acid alignment | http://molevol.cmima.csic.es/castresana/Gblocks_server.html |
| MAFFT | DNA or amino acid sequence alignment | https://mafft.cbrc.jp/alignment/server/ |
| Muscle | DNA or amino acid sequence alignment | https://www.ebi.ac.uk/Tools/msa/muscle/ |
| Prank | DNA or amino acid sequence alignment | http://wasabiapp.org/software/prank/ |
| Probcons | Amino acid sequence alignment | http://probcons.stanford.edu/ |
| T-Coffee | Sequence alignment | http://tcoffee.crg.cat/ |
List of programs for sequence alignment
Phylogenetic analysis
Phylogenetic inference
The evolutionary history of gene, protein or species is generally presented as a phylogenetic tree, a graphical illustration of the evolutionary relationships between the studied taxa. Several methods for phylogenetic inference exist: Maximum Parsimony, the distance-methods and the probabilistic methods. Choose an appropriate method to reconstruct the phylogenetic tree of the genes/proteins of interest. The probabilis| A | | --- | | |
e most widely used for molecular data, but many studies use other methods, distance methods in complement.
Choose one or several of these methods to reconstruct the evolutionary history of your gene, protein or species of interest.
He| A | | --- | | |
reconstruction using diverse methods, and visualization of phylogenetic trees that can be used in complement. For beginners, we recommend using SeaView or Mega with ML analyses, which are very user-friendly and include several tools for sequence alignment, phylogenetic inference including maximum parsimony, distance methods and probabilistic methods, and a tree visualization tool. Advanced users generally prefer the more complex PhyML or RaxML.
| A | B | C |
|---|---|---|
| Software | Features | Link |
| ETEToolkit | Visualization and analysis of phylogenetic trees | http://etetoolkit.org/ |
| FigTree | Graphic software for phylogenetic trees | http://tree.bio.ed.ac.uk/software/figtree/ |
| ITOL | Visualization and annotation of phylogenetic trees | https://itol.embl.de/ |
| Mega | Sequence alignment, model selection, phylogenetic analysis (parsimony, distance methods) | https://www.megasoftware.net/ |
| MrBayes | Bayesian phylogenetic inference, ancestral states reconstruction, phylogenetic calibration… | http://nbisweden.github.io/MrBayes/ |
| PAML | Maximum likelihood phylogenetic inference, estimation of selection strength, ancestral states reconstruction and other analyses | http://abacus.gene.ucl.ac.uk/software/paml.html |
| PAUP | Phylogenetic analyses | http://paup.phylosolutions.com/ |
| PhyML | Maximum likelihood phylogenetic inference, ancestral states reconstruction and other analyses | http://atgc.lirmm.fr/phyml/ |
| RaxML | Maximum likelihood phylogenetic inference | https://cme.h-its.org/exelixis/web/software/raxml/ |
| SeaView | Sequence alignment and phylogenetic analysis (parsimony, NJ, ML) | http://doua.prabi.fr/software/seaview |
List of programs and databases for phylogenetic analysis using diverse methods
| A |
|---|
Option 1: Maximum parsimony
Maximum parsimony is a classic and simple method, that calculates the minimum number of evolutionary steps, including nucleotide insertions, deletions or substitutions, between species.
However, this method ignores hidden mutations and does not take into account branch lengths, potentially leading to long branch attraction, which is an incorrect clustering of unrelated taxa. Furthermore, it does not consider the possibility of hidden mutations, making it not relevant for distant taxa. While maximum parsimony is still used for morphological data, it is no longer considered relevant for molecular data.
Option 2: Distance methods
The Unweighted Pair Group Method with Arithmetic mean (UPGMA), Neighbor Joining (NJ), and Minimum Evolution (ME) are all based on the overall molecular distance, defined by the number of differences between the sequences.
However, this method also ignores hidden mutations and is also subject to long branch attraction. These methods, in particular NJ and ME, are still often used in complement to probabilistic methods in scientific studies.
Option 3: Probabilistic methods
The strength of probabilistic methods is the use of specified models of molecular evolution. Probabilistic methods take into account different mutation rates between sites to avoid mutation saturation. Nowadays, almost all studies of phylogenetic reconstruction use probabilistic methods.
They include Maximum Likelihood (ML) and Bayesian inference (BI). ML calculates the probability of observing the data (in this case, the sequence alignment) under different explicit models of molecular evolution. ML aims to identify the best fit model by exploring multiple combinations of model parameters. BI evaluates the probability of each model of molecular evolution given the data. We recommend using different methods of phylogenetic reconstruction, including ML and BI.
In our example, we used Maximum likelihood to build the phylogenetic tree of the P53 family (including the p73, p63 and p53 proteins of different animal species), and FigTree for a graphical representation of the phylogenetic tree.

Model prediction
Probabilistic methods require a selection of the best model of molecular evolution for the data. Several models of nucleotide evolution exist, differing in their number of parameters like mutation rates between sites. Models of protein evolution also exist. A few programs can be used to calculate a criterion (AIC or BIC) to evaluate the likelihood of different published model for the alignment. Use one of them to select the best fit model for your alignment, which is the model with the lowest AIC or BIC criterion.
If you are using probabilistic methods, use the aligned sequences and a model prediction tool to determine the best model of nucleotide or aminoacid substitution.
Here is a list of tools for selection of models of molecular evolution:
Model selectors are also implemented in software for evolutionary analysis such as Mega.
| A | B | C |
|---|---|---|
| Software | Features | Link |
| ModelTest / JModelTest | Nucleotide substitution model selection | http://evomics.org/resources/software/molecular-evolution-software/modeltest/ |
| ProtTest | Aminoacid substitution model selection | https://github.com/ddarriba/prottest3 |
| SMS | Nucleotide or aminoacid model selection included in PhyML | http://www.atgc-montpellier.fr/sms/ |
List of programs for molecular evolution model selection
Tree rooting
Phylogenetic trees can be unrooted or rooted. Rooting a tree consists in identifying ancestral and derived states. This is useful to study the direction of the evolution and interpret the evolution of the studied taxa.
Diverse methods have been developed to root phylogenetic trees:
-Including outgroups (taxa that do not belong to the studied ingroup but are closely related) in the analysis. It is recommended to choose two outgroups, one being more closely related to the ingroup than to the other outgroup. If outgroups are available, this method can be preferred.
-Midpoint rooting, which corresponds to setting the root at the mid-point of the longest branches.
-Molecular clock rooting, a method that assumes that evolution speed is constant between the sequences. Since evolution speed can vary lot between lineages, this method should be restricted to close species.
Working with phylogenies
Reconstruct the evolution of the gene or protein
Sequence alignments and phylogenetic trees can be used to reconstruct diverse aspects of the evolutionary history of genes, proteins and species, as well as the study the genetic structures within populations. In this last section, we provide a brief and non-exhaustive overview of the main evolutionary studies that can be performed using bioinformatic tools.
Reconstitution of ancestral states
Retracing the functional evolution of genes, proteins, or biological traits often requires the reconstitution of ancestral states. Ancestral states can be inferred from a phylogenetic tree using maximum of parsimony, maximum likelihood, or Bayesian inference; and requires the aligned sequences and the model of molecular evolution that has been used for the phylogenetic analysis (when using probabilistic methods only).
| A | B | C |
|---|---|---|
| Software | Features | Link |
| BayesTraits | Evolutionary analyses using Bayesian inference | http://www.evolution.rdg.ac.uk/BayesTraitsV3.0.5/BayesTraitsV3.0.5.html |
| BEAST | Phylogenetic calibration | http://www.beast.community |
| Mega | Sequence alignment, model selection, phylogenetic analysis (parsimony, distance methods) | https://www.megasoftware.net/ |
| Mesquite | Comparative analyses and statistics | http://www.mesquiteproject.org/ |
| MrBayes | Bayesian phylogenetic inference, ancestral states reconstruction, phylogenetic calibration… | http://nbisweden.github.io/MrBayes/ |
| PAML | Maximum likelihood phylogenetic inference, estimation of selection strength, ancestral states reconstruction and other analyses | http://abacus.gene.ucl.ac.uk/software/paml.html |
| RASP | Ancestral states reconstruction | http://mnh.scu.edu.cn/soft/blog/RASP/index.html |
List of programs and databases for ancestral states reconstruction
| A |
|---|
Measure of selection strength
The strength of selection on protein coding genes can be calculated by evaluating the ration of the number of non-synonymous mutations (mutations changing the protein sequence) per non-synonymous site (dN) and the number of synonymous mutations (mutations with no effect on the protein sequence due to the redundancy of the genetic code) per synonymous site (dS). The ratio dN/dS reveals, if >1, that non-synonymous mutations are higher than expected and the gene is under positive selection. If dN/dS<1, the gene is under purifying selection and if dN/dS=1, the selection is neutral. Selection strength can be calculated by sequence alignment programs such as Mega.
Phylogenetic calibration
Phylogenetic calibration consists in estimating the age of speciation events with events with a known age, i.e., using fossil and other geological data (that can only give minimal ages) as calibration points. Alternatively, mutation rates can be used to calculate the divergence time between two sequences.
Databases such as Timetree, also implemented in Mega, compute the estimated divergence time between species. Mesquite also provides tools to calibrate phylogenetic trees in geological times using fossil data. Ohnologs can also be used to estimate the divergence time between homologues resulting from whole genome duplications in vertebrates.
| A | B | C |
|---|---|---|
| Software | Features | Link |
| BayesTraits | Evolutionary analyses using Bayesian inference | http://www.evolution.rdg.ac.uk/BayesTraitsV3.0.5/BayesTraitsV3.0.5.html |
| BEAST | Phylogenetic calibration | http://www.beast.community |
| Mega | Sequence alignment, model selection, phylogenetic analysis (parsimony, distance methods) | https://www.megasoftware.net/ |
| Mesquite | Comparative analyses and statistics | http://www.mesquiteproject.org/ |
| MrBayes | Bayesian phylogenetic inference, ancestral states reconstruction, phylogenetic calibration… | http://nbisweden.github.io/MrBayes/ |
| Ohnologs | Database of vertebrate ohnologues, resulting from whole genome duplications | http://ohnologs.curie.fr/ |
| Timetree | Tree calibration | http://www.timetree.org/ |
List of programs and databases that can be used for phylogenetic calibration using diverse methods
Study of coevolution
Evolutionary ecology and parasitology often study the evolution of hosts and parasites association through an approach of coevolution, or reciprocal genetic change between different species. Coevolution can also be used to study associations of genes or proteins. Studying coevolution consists in identifying evolutionary events such as co-speciation, host change, and duplication or loss of interaction from the phylogenetic trees of the hosts and the parasites.
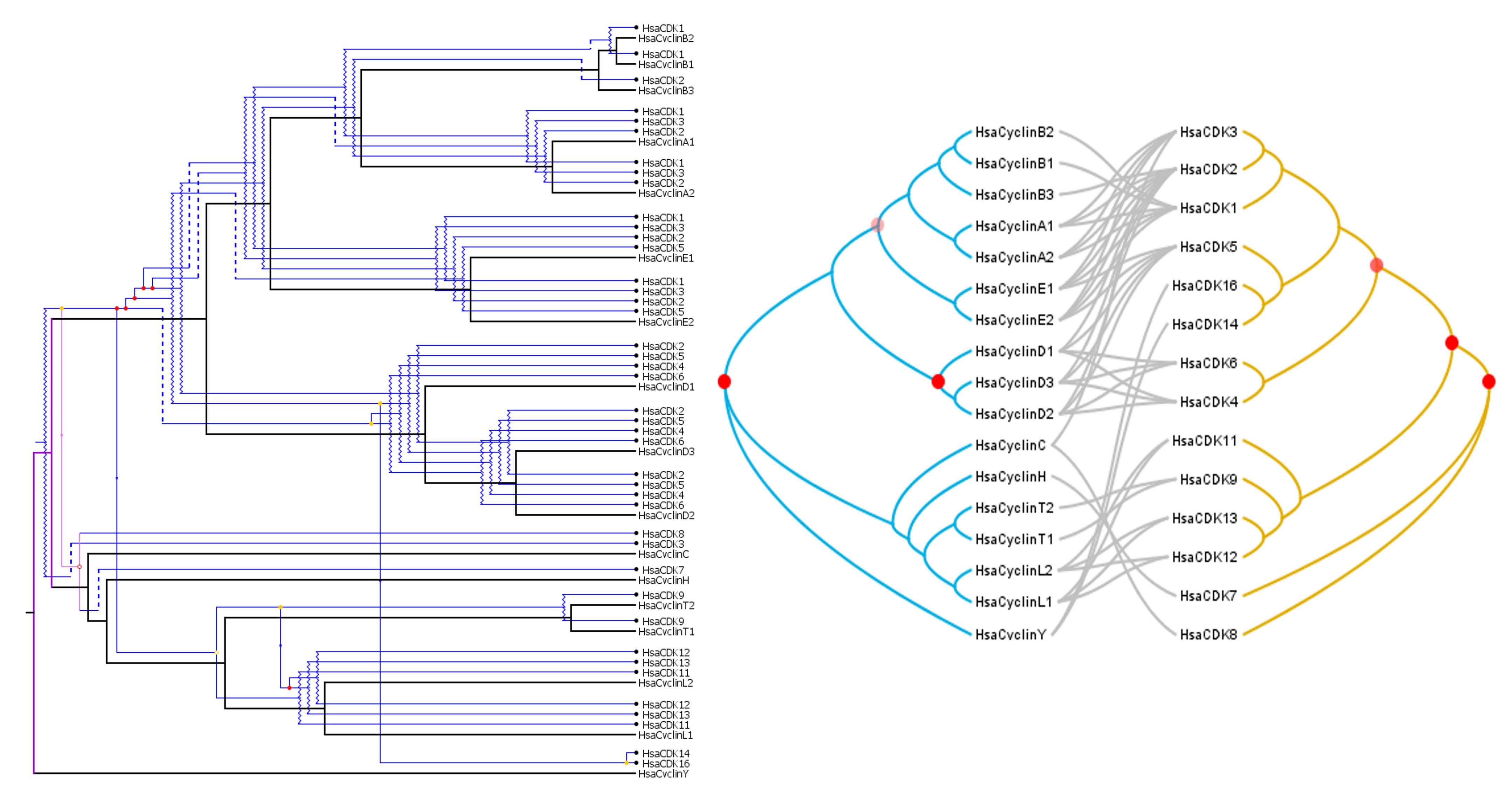
In our second example, we used Maximum likelihood to reconstruct the evolution of cyclins and CDKs, two families of proteins involved in cell cycle control and closely interacting. We used Jane and TreeMap to reconstruct coevolutionary scenarios between the two families.
In both figures, the clustering of cyclins and CDKs indicate an interaction (in this case, that the cyclin can bind the CDK). Red spots indicate significant events of coevolution between the two families of proteins. Co-speciation (hollow red circle), duplications (solid red circle), duplications with host switch (yellow circle), loss of interaction (dashed lines), failures to diverge (jagged lines) are indicated on the cophylogeny.
| A | B | C |
|---|---|---|
| Software | Features | Link |
| Copycat | Software for studying coevolution | http://www.cophylogenetics.com/ |
| Core-PA | Software for studying coevolution | http://pacosy.informatik.uni-leipzig.de/49-1-CoRe-PA.html |
| Jane | Software for studying coevolution | https://www.cs.hmc.edu/~hadas/jane/ |
| TreeMap | Software for studying co-evolution | https://sites.google.com/site/cophylogeny/treemap |
List of programs to study coevolution
Phylogenetic comparative analysis
Evolutionary biology often employs the so-called phylogenetic comparative methods to study the adaptive significance of biological traits. These methods aim at identifying biological characters, in terms of morphology, physiology, or ecology, that result from a shared ancestry. Comparative analyses can be done for quantitative or qualitative variables. A very appropriate tool for comparative analysis and to compute statistics on phylogenetic trees is Mesquite.´BayesTraits can also be used.
Genome evolution
Evolutionary events, such as mutations, insertions, deletions, gene or whole genome duplications, genome reorganization, and genetic exchanges can be identified using phylogenetic trees in complement with genomics tools and databases. Here is a list of databases tools to study diverse aspects of genome evolution, including genome browsers of diverse lineages and tools for comparative genomics and evolutionary genomics:
| A | B | C |
|---|---|---|
| Software | Features | Link |
| BAR | Database of plant genes and proteins | http://bar.utoronto.ca/ |
| CAFE | Gene family evolution | https://github.com/hahnlab/CAFE5 |
| CoGE | Comparative genomics analyses | https://genomevolution.org/coge/ |
| Ensembl | Genome browser of vertebrates, includes tools for identification of homology | https://www.ensembl.org/index.html |
| Entrez | Gene sequences and structures | https://www.ncbi.nlm.nih.gov/Web/Search/entrezfs.html |
| FlyBase | Genome and proteins of the model insect D. melanogaster | https://flybase.org/ |
| GenBank | Annotated DNA sequences | https://www.ncbi.nlm.nih.gov/genbank/ |
| HGT-Finder | Horizontal gene transfer finding | http://cys.bios.niu.edu/HGTFinder/ HGTFinder.tar.gz |
| Ohnologs | Database of vertebrate ohnologues, resulting from whole genome duplications | http://ohnologs.curie.fr/ |
| PomBase | Genes and proteins of the model yeast S. pombe | https://www.pombase.org/ |
| TAIR | Genome and proteins of the model plant A. thaliana | https://www.arabidopsis.org/ |
| WormBase | Genome and proteins of the model nematode C. elegans | https://wormbase.org//#012-34-5 |
| Xenbase | Genome and proteins of the model amphibian X. laevis | http://www.xenbase.org/entry/ |
List of programs and databases to study genome evolution
Population genetics
Genetic diversity can also be explored at the population level by analyzing polymorphism between members of the same species. Gene polymorphisms studies allele diversity within a population, including single nucleotide polymorphisms (SNP), indels, microsatellites or transposable elements. Mathematical models have been developed to describe polymorphism. Several programs are suitable for population genetics studies.
| A | B | C |
|---|---|---|
| Software | Features | Link |
| Arlequin | Population genetics analyses | http://cmpg.unibe.ch/software/arlequin35/ |
| DNAsp | Analysis of DNA polymorphism | http://www.ub.edu/dnasp/ |
| Genepop | Population genetics analyses | https://genepop.curtin.edu.au/ |
| SNiplay | SNP detection and other population genetics analyses | https://sniplay.southgreen.fr/cgi-bin/home.cgi |
List of programs and databases for population genetics
Study of protein structure and function evolution
Studying the functional evolution of proteins can require structure alignments, that can be realized by appropriate programs such as PyMol, and the mean distance in Å between homologous residues can be calculated .
Other programs allow to identify and study structures conservation between proteins and infer a structures from a protein sequence. These analyses and a phylogenetic tree of the protein families, together with the reconstruction of ancestral states, can facilitate the study of the evolution of protein functions within the family.
Here is a list of tools that can be used for analyses on protein structures in an evolutionary framework:
| A | B | C |
|---|---|---|
| Software | Features | Link |
| PyMol | 3D visualization of molecules | http://www.mesquiteproject.org/ |
| I-Tasser | Protein structure prediction | https://zhanglab.dcmb.med.umich.edu/I-TASSER/ |
| Forsa | Protein structure prediction | http://www.bo-protscience.fr/forsa/ |
| HHPred | Protein structure prediction | https://toolkit.tuebingen.mpg.de/tools/hhpred |
List of programs for protein structure analyses

