RT-qPCR Detection of Process Controls (Murine noroviurs and crAssphage) from Wastewater using AB 7500
Jacquelina.Woods, rachel.rodriguez
GenomeTrakr
wastewater
SARS-CoV-2
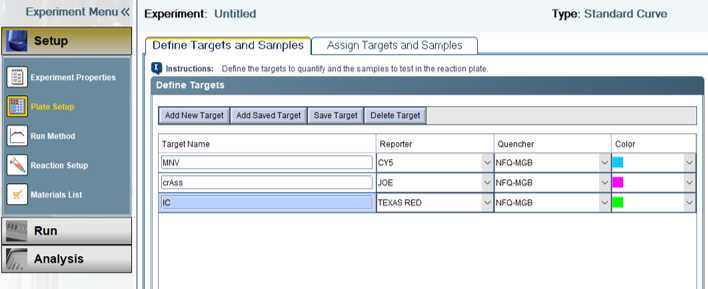
crAssphage
murine norovirus
process control
extraction control
endogenous control
RT-qPCR
AB 7500 Fast
Disclaimer
DISCLAIMER – FOR INFORMATIONAL PURPOSES ONLY; USE AT YOUR OWN RISK
The protocol content here is for informational purposes only and does not constitute legal, medical, clinical, or safety advice, or otherwise; content added to protocols.io is not peer reviewed and may not have undergone a formal approval of any kind. Information presented in this protocol should not substitute for independent professional judgment, advice, diagnosis, or treatment. Any action you take or refrain from taking using or relying upon the information presented here is strictly at your own risk. You agree that neither the Company nor any of the authors, contributors, administrators, or anyone else associated with protocols.io, can be held responsible for your use of the information contained in or linked to this protocol or any of our Sites/Apps and Services.
Abstract
This method was developed at the FDA’s Center for Food Safety and Applied Nutrition for GenomeTrakr’s pandemic response project, monitoring SARS-CoV-2 variants in wastewater. Protocols developed for this project cover wastewater collection, concentration, RNA extraction, RT-qPCR detection, library prep, genome sequencing, quality control checks, and data submission to NCBI. This protocol describes the murine norovirus (MNV; extraction control) and crAssphage (human indicator) RT-qPCR assay developed for use on the AB 7500 platform using software version 2.0 or 2.3. The assay incorporates an internal amplification control (IC) to prevent the reporting of false negatives due to inhibition or failure of the RT-qPCR. This multiplexed detection assay was developed for the determination crAssphage extracted from wastewater, as an endogenous control, and MNV as an extraction control. The assay is designed to be used in conjunction with the SARS-CoV-2 RT-qPCR detection assay. Valid sample results for SARS-CoV-2 detection are contingent upon the detection of the MNV extraction control from the sample being tested.
Before start
Always wear gloves during this procedure and never wear the same gloves when going between master mix and samples.
Always use aerosol resistant pipette tips for PCR.
Steps
Master Mix Preparation
Prepare Master Mix for all sample and control reactions based on the volumes per 25 µl reaction in this table. Composition of mixes are listed here: Reagent Mixes for RT-qPCR Detection of Process Controls from Wastewater Samples (protocols.io) and should be prepared in advance and stored appropriately. Alternatively, Master Mixes can be prepared from individual components as described here: Master Mix Table for MNV-crAss Assay.pdf.
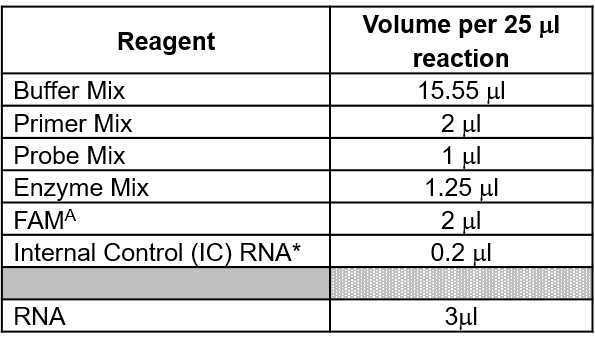
Thaw Master Mix reagents in bench top cool block (chilled at 2-8°C) or 4On ice in master mix preparation hood.
Vortex reagent tubes for 0h 0m 3sat setting medium high to high (if vortex has settings).
Briefly centrifuge all reagents 0h 0m 5s in a personal microcentrifuge to bring liquid to the bottom of tube.
Return all reagents to bench top cool block (chilled at 2-8°C) or 4On ice.
Proceed to hood/area or room where the template is added and thaw IC RNA and sample RNA in this hood/area.
Briefly centrifuge IC RNA 0h 0m 5s in a personal microcentrifuge to bring liquid to the bottom of tube.
Add appropriate volume of IC RNA (0.2µLper reaction) to Master Mix from Step 1.4 in cold block/on ice.
Vortex briefly and centrifuge 0h 0m 5s in a personal microcentrifuge.
Reaction Set-Up
Add 22µL of Master Mix to each designated reaction tube or sample wells.
Briefly centrifuge sample RNA 0h 0m 5s in a personal microcentrifuge to bring liquid to the bottom of tube.
Add 3µL of sample RNA template to each of three reaction tubes or wells.
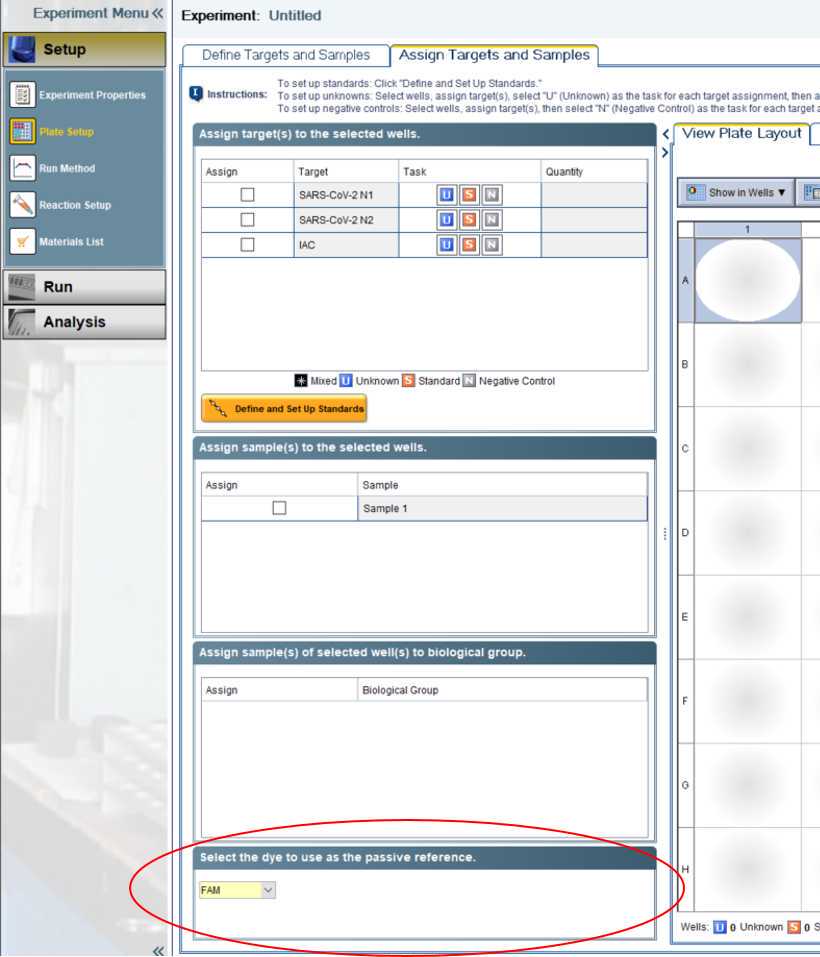
Ensure each plate or run has appropriate controls (positive and negative controls) included.
Seal sample plate or strip tubes. Then, briefly spin 0h 0m 5s.
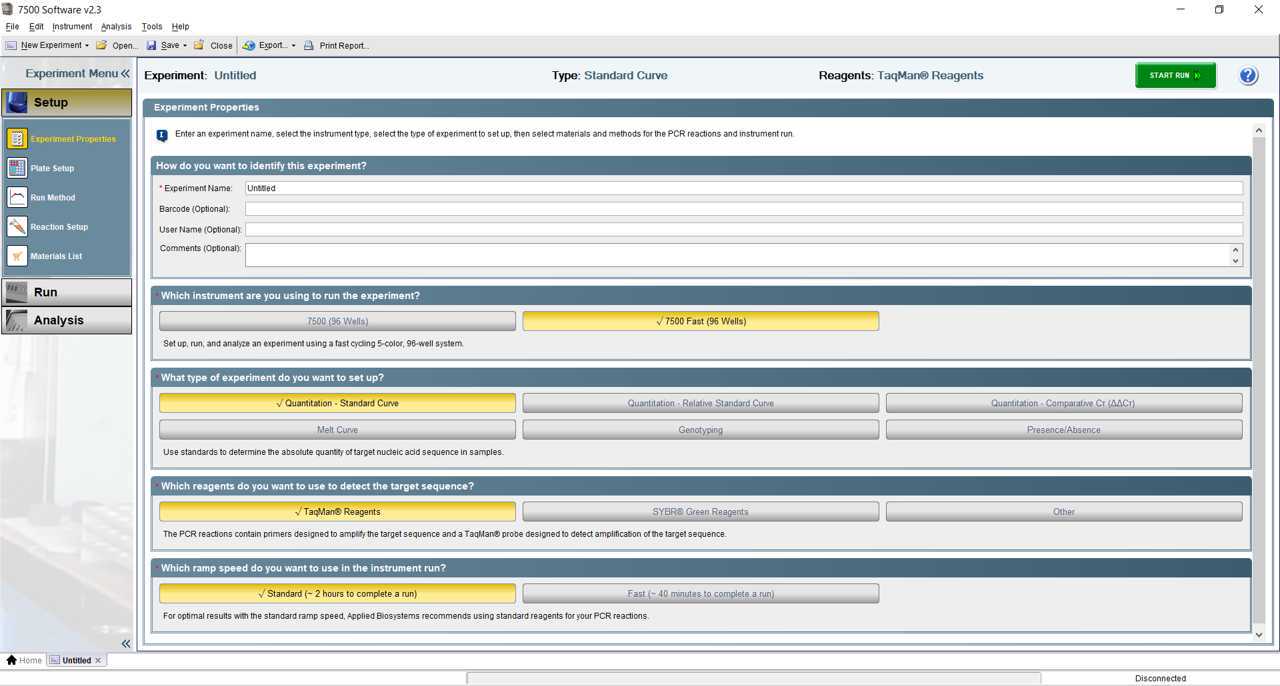
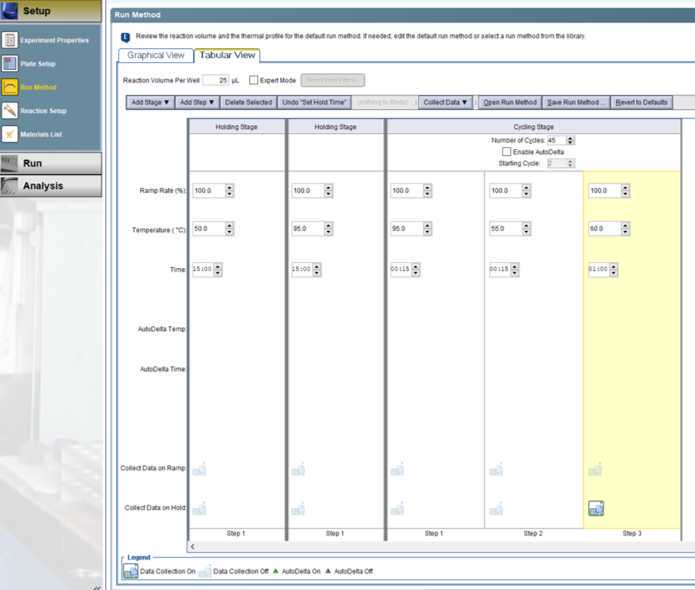
Start run on Applied Biosystems 7500 Fast instrument.
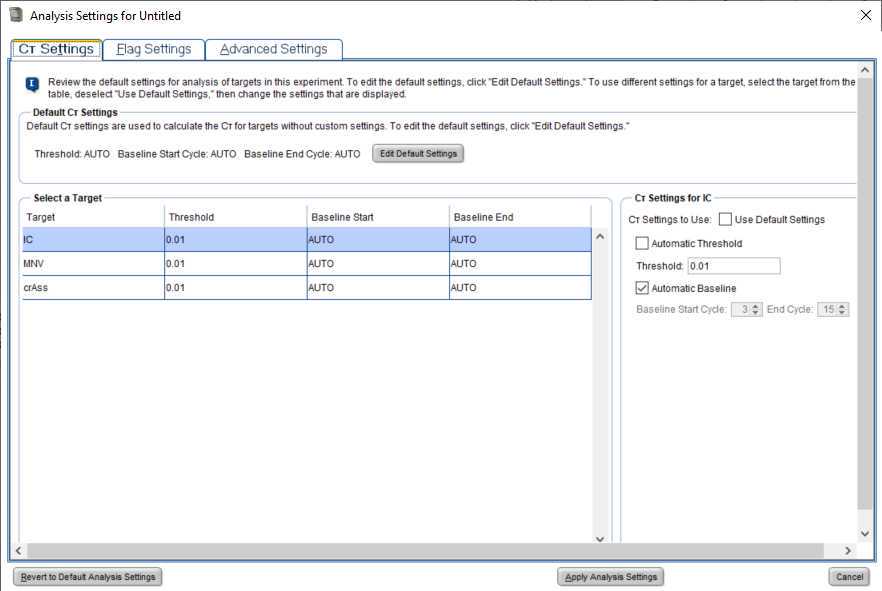
Data Analysis
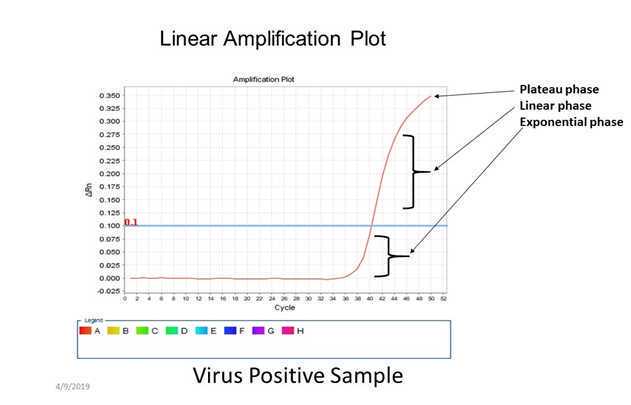
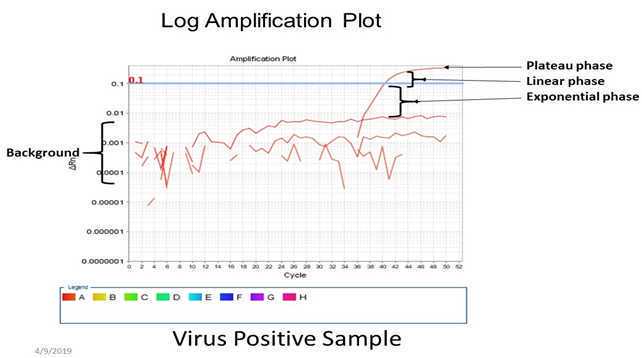
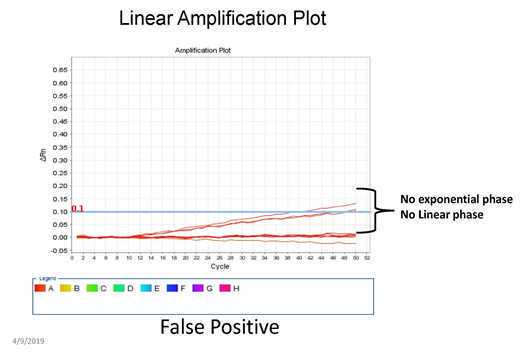
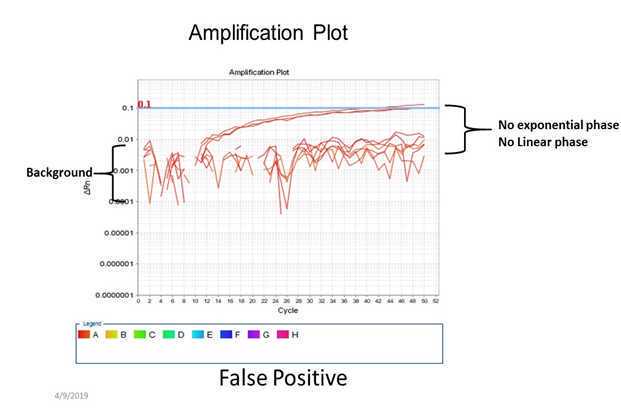
Verify positive and negative calls for each reaction using either linear or log amplification plots.
Sample is invalid if any of the following are observed:
-
Negative RT-qPCR control is positive (Ct value indicated) for any of the expected targets (MNV or crAssphage);
-
Positive RT-qPCR control is negative (undetermined) for expected target/s (MNV or crAssphage);
-
Sample is negative (undetermined) for MNV; or
-
IC is negative (undetermined) in the sample, or the average of the IC Ct values from the sample replicates are greater than 4 Cts than the IC Ct of the negative RT-qPCR control.
Sample is valid if all of the following are observed:
-
Negative RT-qPCR control is negative (undetermined) for expected target/s (i.e., MNV and crAssphage);
-
Positive RT-qPCR control is positive (Ct value indicated) for expected target/s (i.e., MNV and crAssphage); and
-
Internal amplification control (IC) is positive in all sample reactions with the average IC Ct values for the sample is less than 4 Cts greater than the IC Ct of the negative RT-qPCR control.