Quant-iT™ PicoGreen® dsDNA Quantification
Roey Angel, Eva Petrova
Abstract
The following protocol is intended for the quantification of double-stranded DNA using Quant-iT™PicoGreen® dsDNA Assay Kit (ThermoFisher® dsDNA Assay Kit (ThermoFisher). This protocol is a simplified and condensed version of the full protocol from the manufacturer. The procedure described here is for 96 reactions. If samples are run in duplicates, then this should allow quantifying 40 samples.
Before start
This protocol is optimised for measuring an entire 96-well plate. It assumes that 16 wells will be used for measuring the standards and the blank samples (in duplicates) and 80 wells will be used for measuring unknown DNA samples (typically in duplicates).1. The protocol can be easily adjusted for a lower number of samples by reducing the volume of the working solutions of the reagents. Note though that enough TE should be retained for diluting the standard stock solution (490 or 680 µl), for potentially diluting the unknown samples, if their concentration is too high, and for accounting for pipetting errors. To fill the plate, 19.2 ml of TE is needed. So if only 40 wells are to be used for measuring unknown samples prepare about
- The dynamic range of the assay is between 50 pg ml-1 to 1000 ng ml-1. This translates into DNA sample concentrations of 0.05-5 ng µl-1 and 1-200 ng µl-1 in the low-range and high-range assays, respectively. Samples with higher DNA concentration need to be diluted (e.g. in DNase-free water or TE buffer).
- Note that some compounds that can be present as DNA contaminations (e.g. salts, ethanol, detergents, proteins) are claimed by the manufacturer to not interfere with the measurement. Please refer to the full protocol for a list of these compounds and their effect on the measurement. Also, equimolar presence of ssDNA and RNA in the sample should have only minimal effect on the quantitation results.
Attachments
Steps
Prepare reaction
Take out all reagents from the fridge and bring them to room temperature.
Take out the DNA samples from the freezer. DNA samples should be slowly thawed on ice.
Prepare 22 ml 1X TE buffer by pipetting 1.1 ml of 20X TE buffer into 20.9 ml of nuclease-free water into a sterile and nuclease-free 50 ml tube.
Mix by inverting the tube several times.
1.1
20.9
For high-range quantification:
Dilute the DNA-standard stock solution (λ DNA 100 ng µl-1) to a final concentration of 2 ng µl-1by mixing 10 µl λ DNA-standard stock solution with 490 µl 1X TE buffer.
10
490
For low-range quantification:
Prepare a 40-fold dilution of the 2 ng µl-1 DNA-standard work solution by mixing 5 μl of the 2 ng μl-1 DNA-standard work solution with 195 μl 1X TE buffer to yield a 0.05 ng µl-1 DNA-standard work solution.
5µL
195µL
If needed, prepare a dilution of each sample in 1X TE buffer so that the reading will be within the dynamic range.
Prepare PicoGreen® work solution: 9950 µL 1X TE buffer + 50 µL PicoGreen®into a sterile and nucleic-acids free 50 ml tube. Mix and protect from light.
9950
50
Prepare the following standard mixture in the first two columns of the black, sterile, 96-well plate:
| A | B | C | D |
|---|---|---|---|
| Assay version | Diluted DNA std. (µl) | 1X TE buffer (µl) | Final DNA amount (ng) |
| High-range (1-200 ng µl-1) | 100 | 0 | 200 |
| Use 2 ng μl-1 standard | 50 | 50 | 100 |
| 10 | 90 | 20 | |
| 1 | 99 | 1 | |
| 0 | 100 | 0 | |
| Low-range (50 pg µl-1 - 5 ng µl-1) | 100 | 0 | 5 |
| Use 0.05 ng μl-1 standard | 50 | 50 | 2.5 |
| 10 | 90 | 0.5 | |
| 1 | 99 | 0.05 | |
| 0 | 100 | 0 |
Equipment
| Value | Label |
|---|---|
| 96-well microtiter plate | NAME |
| Nunc | BRAND |
| 265301 | SKU |
| black, flat bottom | SPECIFICATIONS |
Pipette 99 µl of 1X TE buffer in the remaining wells.
99
Equipment
| Value | Label |
|---|---|
| Multipette E3 | NAME |
| Eppendorf | BRAND |
| 4987000010 | SKU |
| electronic dispenser | SPECIFICATIONS |
Pipette 1 µl of the unknown DNA samples in the remaining wells.
1µL
Pipette 100 µl of the PicoGreen® work solution in each well, including the standard and unknown sample wells.
100
Protect the 96-well plate from light and incubate for 2-5 min at room temperature.
0h 2m 0s
Measure samples
Place the plate in a plate reader and measure the fluorescence according to the following parameters:
Excitation ~480 nm
Emission ~520 nm
Integration time 40 s
Lag time 0 s
Gain Optimal
Number of flashes 10
Calculated well highest standard
Shaking 5 s
Equipment
| Value | Label |
|---|---|
| Synergy 2 | NAME |
| absorbance microplate reader | TYPE |
| BioTek | BRAND |
| Synergy2 | SKU |
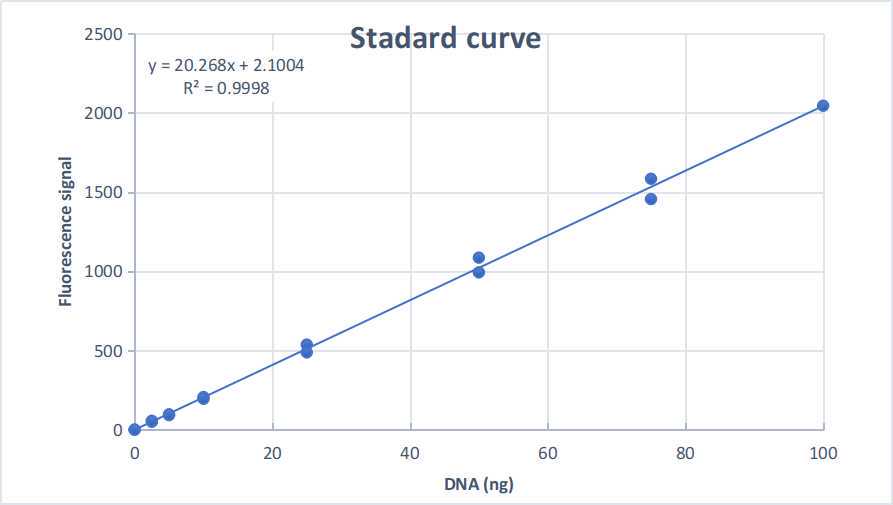
Plot the measured fluorescent values of the standard samples against their known concentrations and fit a linear curve using linear regression. Make sure that the coefficient of determination (R2) is close to 1 (typically > 0.99). Calculate the DNA concentrations in the unknown samples using the slope and intercept parameters of the linear equation. Output values you obtained are in ng µl-1, assuming 1 µl of each sample was used.
See attached example spreadsheet.