Protocol for the Experimental and Bioinformatic Analysis for Gut Microbiota Profiling of Litopenaeus vannamei
Luis Enrique Vazquez-Morado, Melany Cervantes-Echeverría, Fernanda Cornejo-Granados, Luigui Gallardo-Becerra, Filiberto Sánchez-López, Adrian Ochoa-Leyva
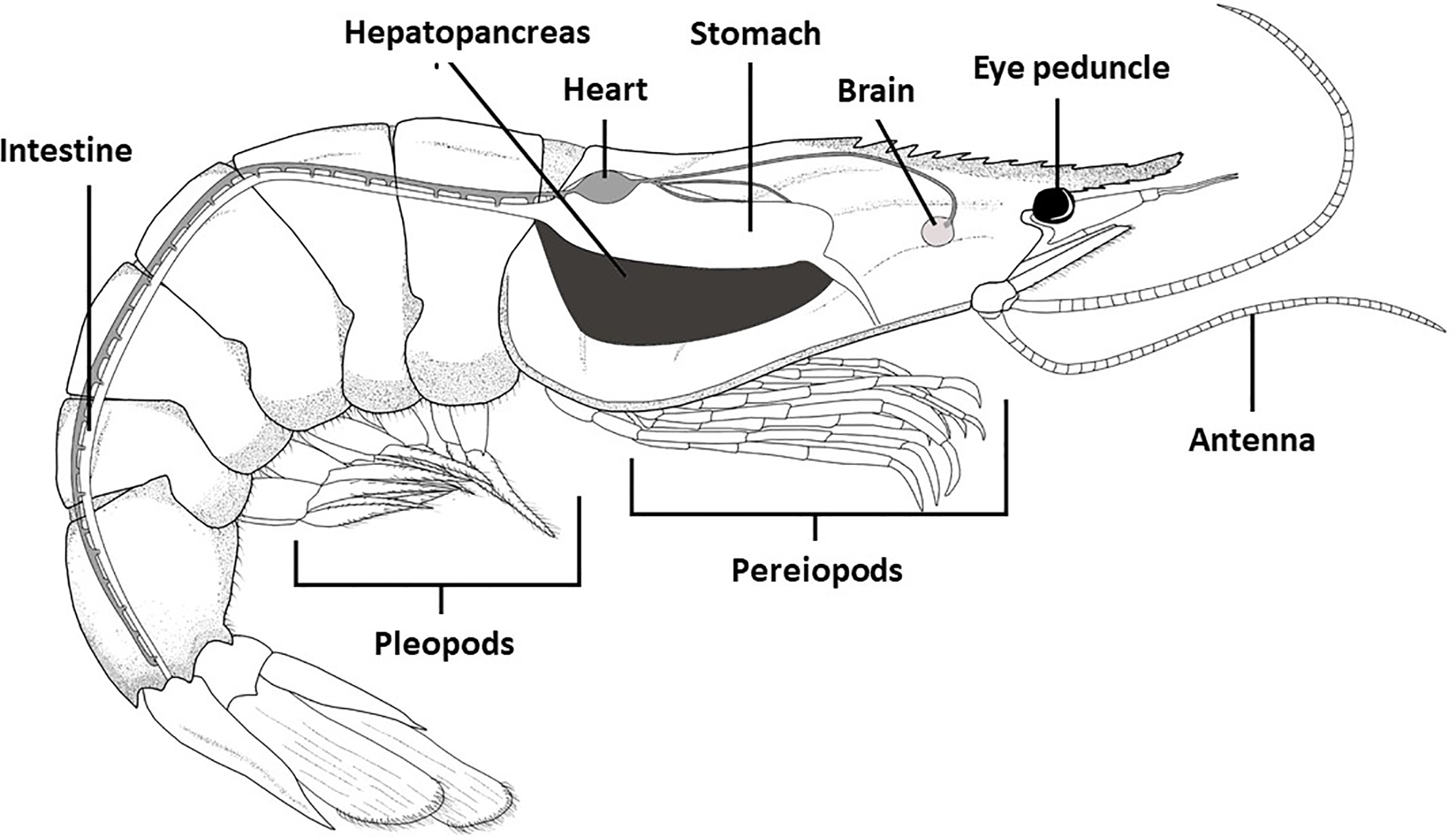
Shrimp Gut Microbiome
DNA Extraction
Microbiome Bionformatics
Shrimp Gut Extraction
16s Gene Amplification
Amplicon Purification
Abstract
The Pacific white shrimp ( Litopenaeus vannamei, Lv ) is a non-model organism of significant global economic importance, constituting 51.7% of crustaceans produced through aquaculture. Consequently, scientific research has focused on various approaches to enhance the production of this crustacean, such as understanding the dynamics of its microbiota. Microbiota refers to the collection of microorganisms inhabiting a host organism, which can be essential for the host's healthy development. The gut is one of the most critical organs for shrimp development and survival. The current protocol outlines the methodology for adequately extracting gut samples from Lv. It describes the DNA extraction method, amplification, and purification of the V3 region of the 16S ribosomal gene and the construction of libraries for subsequent sequencing. Furthermore, it also includes a bioinformatic protocol for sequence analysis to describe the microbiota composition.
Before start
Before starting, it is necessary to have a thoroughly disinfected workspace. Use 10% sodium hypochlorite solutions, 70% ethanol, distilled water, and absorbent paper towels to clean the area. It is advisable to collect intestine samples from shrimp that have been recently taken out of the pond. Otherwise, it is essential to verify the integrity of this organ and confirm that it is not undergoing lysis. Additionally, a basic understanding of Linux and related working environments is recommended for processing the final reads.
Steps
Shrimp Gut Sample Extraction
Before beginning any sample collection, it is crucial to prepare the working environment properly. Start by rinsing the surgical forceps with a 10% chlorine solution, then rinse thoroughly with sterile Milli-Q water. Next, place 1.5 ml Eppendorf tubes with 1 mL of RNAlater or 20% glycerol as preservation reagent in a rack for easy access during sampling. If collecting samples outside the laboratory, be sure to have a container of liquid nitrogen on hand to freeze the samples of 20% glycerol and a cooler with ice to keep the samples with RNAlater for transport. By taking these simple precautions, you can ensure the integrity of your samples and obtain accurate results.
First, to collect a shrimp sample, put on sterile latex gloves and remove the exoskeleton without detaching the cephalothorax zone. Then, hold the shrimp with your dominant hand and apply measured pressure to expose the hepatopancreas and gut. Next, cut along the shrimp following the same line as the gut, careful not to damage the organ. Cut the gut connection with the hepatopancreas, and open the shrimp like a butterfly following the previously made cut. Finally, transfer the gut using surgical forceps to a 1.5 mL Eppendorf tube with preservation reagent. Keep the RNAlater sample at 4ºC for 48 hours and posteriorly maintain it at -70 C until use. Contrary, save the glycerol sample directly in liquid nitrogen and keep it at -70 until use. Remember to take the necessary precautions to ensure the integrity of your samples and obtain accurate results.
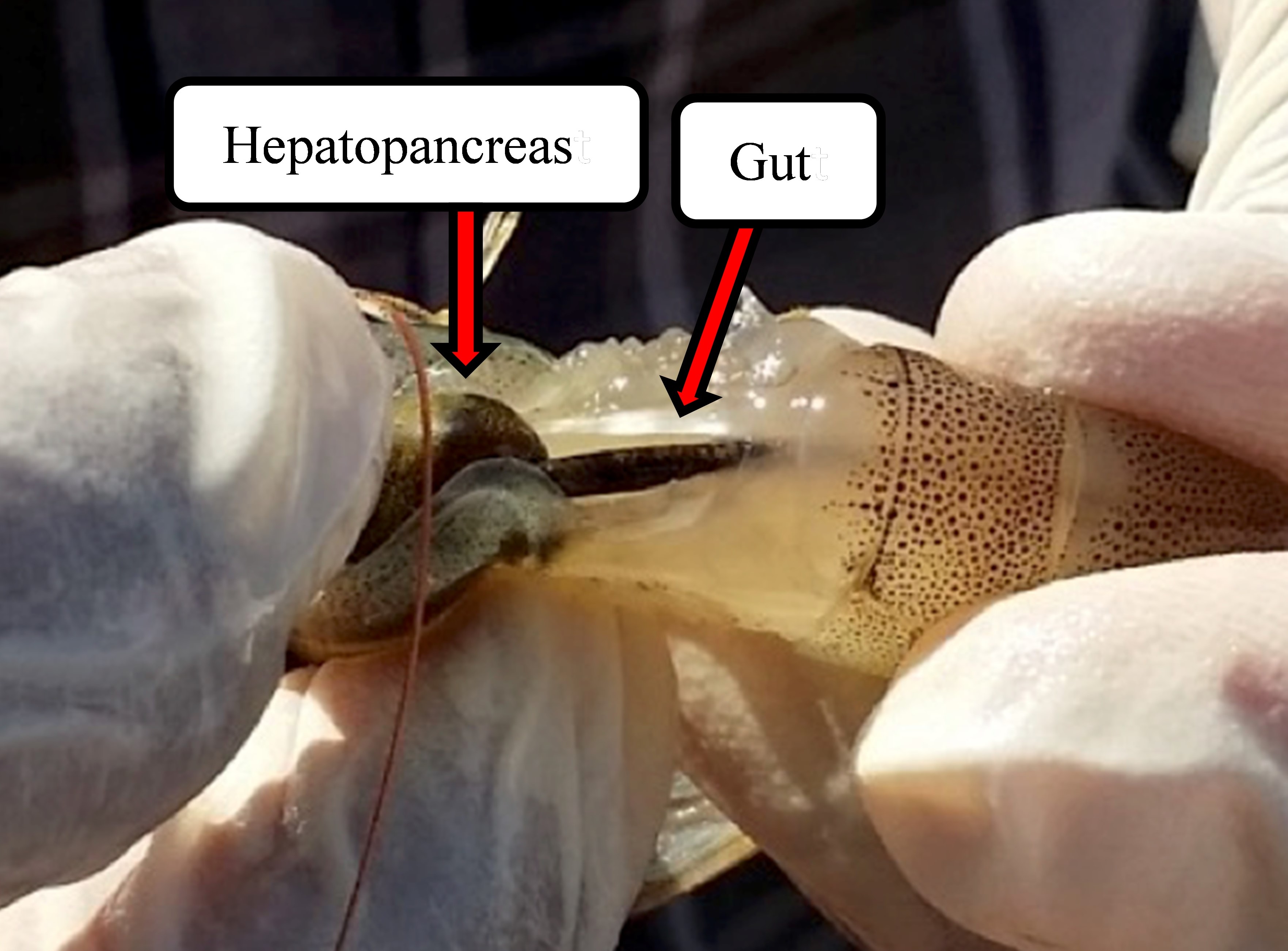
DNA Extraction from Shrimp Gut Samples
Take the previously collected sample from the freezer (-70º C) and let it thaw on ice to extract the genomic DNA. Once the sample is thawed, centrifuge the tube 8,000 g for 5 min. Then, discard the supernatant, add 1 ml of phosphate buffer at pH 7.4, and let it sit on ice for 0.5-1 minute. This is to remove as much of the conservation reagent as possible.
From now on, use the genomic DNA extraction kit Quick-DNA Fecal/Soil Microbe MiniPrep Zymo Research. You can opt to use the ZR Bashing Bead Lysis Tube (mechanical lysis) or use the Genomic Lysis Buffer directly (chemical lysis).
To only use the chemical lysis skip to step 10 and add 1,200 µl of the Genomic Lysis Buffer to the sample. Continue with the following steps as specified.
For the mechanical lysis transfer the gut sample in PBS buffer to a ZR Bashing Bead Lysis Tube and add 700 µl of BashingBead Buffer. Secure the tube in a Vortex Genie2 and agitate for 5 min at maximum speed.
Centrifuge the ZR BashingBead™ Lysis Tube in a microcentrifuge at ≥ 10,000 x g for 1 minute.
Transfer up to 400 µl supernatant to a Zymo-Spin™ III-F Filter in a collection tube and centrifuge at 8,000 x g for 1 minute.
Add 1,200 µl of the Genomic Lysis Buffer to the filtrate in the collection Tube from the previous step. Mix well.
Transfer 800 µl of the mixture from the previous step to a Zymo-Spin™ IICR column in a collection tube and centrifuge at 10,000 x g for 1 minute.
Discard the flow through from the collection tube and repeat the previous step.
Add 200 µl DNA Pre-wash buffer to the Zymo-Spin™ IICR Column in a new collection tube and centrifuge at 10,000 x g for 1 minute
Add 500 µl g-DNA wash buffer to the Zymo-Spin™ IICR Column and centrifuge at 10,000 x g for 1 minute.
Transfer the Zymo-Spin™ IICR column to a clean 1.5 ml microcentrifuge tube and add 100 µl (50 µl minimum) DNA elution buffer directly to the column matrix. Centrifuge at 10,000 x g for 30 seconds to elute the DNA.
Place a Zymo-Spin™ III-HRC filter in a clean collection Tube and add 600 µl prep solution. Centrifuge at 8,000 x g for 3 minutes.
Transfer the eluted DNA to a prepared Zymo-Spin™ III-HRC Filter in a clean 1.5 ml microcentrifuge tube and centrifuge at exactly 16,000 x g for 3 minutes.
The purified DNA should be stored at -20ºC until use. For long storage periods ranging from months to years, -80 °C is recommended.
To quantify DNA recovered in previous extraction, use a fluorometric method as Qubit dsDNA HS Assay with the manufacturer's recommended conditions.
Manual:
It is necessary to assess the quality of the extracted DNA extracted by agarose gel (2%) electrophoresis using fluorescent staining of dsDNA. An appropriate DNA size marker should be loaded along with the samples. The gel should be run at a constant voltage of 100 volts for 30 minutes. Finally, to visualize the gel in UV-transilluminator.
Amplification and Purification of the V3 region of the 16s Ribosomal Gene
Prepare the following PCR reaction using the DNA extracted from shrimp gut:
V-Add the required amount of sample and make up to 20 µl with water.
| A | B |
|---|---|
| DNA Template (~12.5 ng) | V |
| Forward primer rRNA 16s Gene (10 pmol/µl) | 1 |
| Reverse primer rRNA 16s Gene (10 pmol/µl) | 1 |
| Reagent Q5 Hot Start Master Mix | 10 |
| H2O | x |
| Final Volume | 20 |
Preparation of the PCR Reaction for the Amplification of Regions of the 16s Ribosomal Gene
Amplify under the following conditions in the thermocycler:
A: Temperature (°C)
B: Time
C: Cycles
| A | B | C | D | E |
|---|---|---|---|---|
| A | Initial denaturation | 98 | 1 m | 1 |
| B | Denaturation | 98 | 30 s | 30 |
| B | Annealing | 55 | 30 s | 30 |
| B | Extension | 72 | 30 s | 30 |
| C | Final extension | 72 | 5 m | 1 |
| D | Hold | 4 | ∞ | 1 |
Thermalcycler Conditions
In this step, purify the amplicons using AMPure XP beads. Before beginning, bring the beads to room temperature with at least one hour before use, prepare 80% ethanol solution and keep it refrigerated at -20 °C.
Transfer the entire Amplicon PCR product to a 1.5 ml Eppendorf tube and add nuclease-free water until reaching a final volume of 100 µl.
Vortex the AMPure XP beads to homogenize them. Add 50 μl of AMPure XP beads to each sample tube. Gently homogenize the mixture by vortexing at a low intensity, and then incubate it at room temperature for 5 minutes.
Place the tube with the sample on a DynaMag™-2 magnetic rack until the mixture clears. Then, recover the supernatant by transferring it to a new 1.5 mL Eppendorf tube.
Add 15 µL of Ampure XP beads to the obtained supernatant. Then, homogenize in vortex for 1 min at low intensity and incubate the sample at room temperature for 5 minutes.
Place the tube with the sample in the magnetic rack until it clears and discard the supernatant.
While the tube is on the magnetic rack, add 200 μl of 80% ethanol to wash the beads without homogenizing. Remove the ethanol carefully and repeat this step once more for a total of two wash steps.
Maintain the tube with the sample on the magnetic rack and allow to air-dry for a maximum of 10 min until the ethanol evaporates.
Add 22 µl of nuclease-free water to the tube, homogenize, and incubate at room temperature for 5 minutes.
Place the tube with the sample in the magnetic rack until it clears and recover the supernatant. Store the purified amplicon at -80 °C until use.
Verify the integrity of the amplicon in a 2% agarose gel and quantify the final DNA obtained.
V3 Metagenomic Library Preparation.
Index addition
The next step is to attach dual indices and Illumina sequencing adapters using the Nextera XT Index Kit
1- Transfer 5 μl from purified amplicon to a new Eppendorf tube.
Add 5 μl Index 1 to the tube with purified amplicon.
Add 5 μl Index 2 to the tube with purified amplicon.
Set up the following reaction of DNA:
| A | B |
|---|---|
| Reagent Q5 Hot Start Master Mix 2X | 25 μl |
| PCR Grade Water | 25 μl |
| Nextera XT Index 1 Primers (N7XX) | 5 μl |
| Nextera XT Index 2 Primers (S5XX) | 5 μl |
| DNA template | 5 μl |
| Final Volume | 50 μl |
Gently pipette up and down 10 times to mix.
Perform PCR on a thermal cycler using the following program:
| A | B | C | D | E |
|---|---|---|---|---|
| A | Initial denaturation | 98 | 1 m | 1 |
| B | Denaturation | 98 | 30 s | 12 |
| B | Annealing | 55 | 30 s | |
| B | Extension | 72 | 30 s | |
| C | Final extension | 72 | 5 m | 1 |
| D | Hold | 4 | ∞ | 1 |
PCR Clean-Up
This step uses AMPure XP beads to clean up the final library before quantification.
Centrifuge the Index PCR tube at 280 x g at 20 °C for 1 minute to collect condensation.
Vortex the AMPure XP beads for 30 seconds to make sure that the beads are evenly dispersed. Add an appropriate volume of beads to a trough.
Add 56 μl of AMPure XP beads to the Index PCR tube.
Gently pipette mix up and down 10 times.
Incubate at room temperature without shaking for 5 minutes.
Place the tube on a magnetic stand for 2 minutes, or until the supernatant has cleared.
With the Index PCR tube on the magnetic stand, use a pipette to remove and discard the supernatant.
With the Index PCR tube on the magnetic stand, wash the beads with freshly prepared 80% ethanol as follows:
-Using a multichannel pipette, add 200 μl of freshly prepared 80% ethanol to the tube.
-Incubate the tube on the magnetic stand for 30 seconds.
-Carefully remove and discard the supernatant.
With the Index PCR tube on the magnetic stand, perform a second ethanol wash as follows:
-Using a multichannel pipette, add 200 μl of freshly prepared 80% ethanol to the tube.
-Incubate the tube on the magnetic stand for 30 seconds.
-Carefully remove and discard the supernatant.
-Use a pipette with fine pipette tips to remove excess ethanol.
With the Index PCR tube still on the magnetic stand, allow the beads to air-dry for 10 minutes.
Remove the Index PCR tube from the magnetic stand.
Using a pipette, add 27.5 μl of H2O to the Index PCR tube.
Incubate at room temperature for 2 minutes.
Place the tube on the magnetic stand for 2 minutes, or until the supernatant has cleared.
Using a pipette, carefully transfer 25 μl of the supernatant from the Index PCR tube to a new eppendorff tube.
Library Quantification
Illumina recommends quantifying your libraries using a fluorometric quantification method that uses dsDNA binding dyes.
Calculate DNA concentration in nM, based on the size of DNA amplicons as determined by an Agilent Technologies 2100 Bioanalyzer trace:
(concentration in ng/μl) × 106 = (660 g/mol × average library size)
For example:
15 ng/μl × 106 = (660 g/mol × 500)
concentration in nM
45 nM
Sequencing the sample in 2x150 pb for the V3 region is necessary, with a recommended total number of reads of 100k to 200k per sample. The amount of DNA recommended in this protocol may vary between service units.
Analysis of the shrimp microbiome using DADA2 by Qiime2
After sequencing, data files in a FASTQ format containing the nucleotide sequences or reads are generated. A computer or server with Ubuntu 22.04 operating system or some other linux distribution to the user's taste should be used for the analysis of the reads. The sequences can be worked in the same way in macOS terminals or in Ubuntu terminals installed in windows with the WSL options enabled.
Before starting, create a directory dedicated solely for the analysis of the reads.
$ mkdir workspace
Subsequently, enter the directory created and save the raw reads.
$ cd workspace
$ mkdir raw_seqs
Raw reads should be stored in the ¨raw_seqs¨ folder.
Perform a quality visualization of the reads from FASTQ file with fastQC.
$ fastqc Archive_R1.fastq Archive_R2.fastq
or
$ fastqc Archive*fastq
Remove NEXTERA adapters with Cutadapt.
$ cutadapt -b "file:adapter_list.fasta" -o Archive_no_adapters_R1.fastq Archive_R1.fastq
$ cutadapt -b "file:adapter_list.fasta" -o Archive_no_adapters_R2.fastq Archive_R2.fastq
The file with the Nextera adapters list (adapter_list.fasta) needs to be created with the following sequences:
PrefixNX/1
AGATGTGTATAAGAGACAG
PrefixNX/2
AGATGTGTATAAGAGACAG
Trans1
TCGTCGGCAGCGTCAGATGTGTATAAGAGACAG
Trans1_rc
CTGTCTCTTATACACATCTGACGCTGCCGACGA
Trans2
GTCTCGTGGGCTCGGAGATGTGTATAAGAGACAG
Trans2_rc
CTGTCTCTTATACACATCTCCGAGCCCACGAGAC
Use the Trimmomatic tool to quality-trim your reads. Before selecting any program option, carefully review the characteristics of your reads in the fastQC results. The files can be used in a compressed .gz format.
$ TrimmomaticPE -phred33 -threads 8 Archive_no_adapters_R1.fastq Archive_no_adapters_R2.fastq Archive_trimmo_results_R1.fastq unpaired_reads_R1.fastq Archive_trimmo_results_R2.fastq unpaired_reads_R2.fastq OPTION1:
The resulting unpaired files will not be used in the following steps, only the "trimmo" paired files.
Once your sequences have been quality-trimmed, modify the file names of your reads according to the following structure (this is mandatory):
Name Structure:
1 2 3 4 5 6
SampleName_AAAAAAAA_L001_R1_001.fastq
1- Name of the sample.
2- 8-letter code that must be unique for each sample (it must be the same for the R1 and R2 pair).
3- This should be as shown for all sequences.
4- Sequence R1 or R2.
5- This should be as shown for all sequences.
6- The format in fastq.
Then the sequences should be compressed to "gz" format.
Create a metadata.txt file in a text table format that includes the sample names, labels, sample origins, and any comparisons you need to develop. Here's an example template you can use:
| A | B | C | D | E |
|---|---|---|---|---|
| #SampleID | BarcodeSequence | Group | Treatment | Organ |
| sample_01 | AAAAAAAA | 1 | 1 | Gut |
| sample_02 | AAAAAAAC | 2 | 2 | Gut |
| sample_03 | AAAAAAAG | 3 | 3 | Gut |
Metadata Structure
Now, let's start using the Qiime2 tool.
Activate the conda environment of qiime2:
$ conda activate qiime2-2023.5
Import the reads to the qiime2 program:
$ qiime tools import --type 'SampleData[PairedEndSequencesWithQuality]' --input-path quality_reads/ --input-format CasavaOneEightSingleLanePerSampleDirFmt --output-path work_space/demux-paired-end.qza
Performs the process of denoising the reads to obtain the ASVs:
$ cd work_space/
$ qiime dada2 denoise-paired --i-demultiplexed-seqs demux-paired-end.qza --p-trunc-len-f 0 --p-trunc-len-r 0 --o-table PE-table.qza --o-representative-sequences PE-rep-seqs.qza --o-denoising-stats PE-stats.qza
Converts the resulting files to "qzv" format, which can be directly viewed on the page https://view.qiime2.org
$ qiime metadata tabulate --m-input-file PE-stats.qza --o-visualization PE-stats.qzv
$ qiime feature-table summarize --i-table PE-table.qza --o-visualization PE-table.qzv --m-sample-metadata-file metadata.txt
$ qiime feature-table tabulate-seqs --i-data PE-rep-seqs.qza --o-visualization PE-rep-seqs.qzv
From here on, we will proceed with the taxonomic annotation of your ASVs.
Train a classifier for the 16S ribosomal gene. Obtain the necessary sequences directly from the SilvaDB website. We recommend using the nr99_ref sequences, always in their latest version.
$ qiime feature-classifier fit-classifier-naive-bayes --i-reference-reads silva_version_nr99_ref_seqs.qza --i-reference-taxonomy silva-version-99-tax.qza --o-classifier silva_version_nr99_classifier.qza
Perform the annotation of your ASVs with the following commands:
$ qiime feature-classifier classify-sklearn --i-classifier silva_version_nr99_classifier.qza --i-reads PE-rep-seqs.qza --o-classification PE-taxonomy_silva_version.qza
$ qiime taxa collapse --i-table PE-table.qza --i-taxonomy PE-taxonomy_silva_version.qza --p-level 7 --o-collapsed-table table_taxa_PE.qza
$ qiime taxa barplot --i-table PE-table.qza --i-taxonomy PE-taxonomy_silva_version.qza --m-metadata-file metadata.txt --o-visualization PE-taxa-bar-plots_silva_version.qzv
From this step onwards, we will begin calculating alpha and beta diversity.
Create a rooted tree of the ASVs using the following commands:
$ qiime alignment mafft --i-sequences PE-rep-seqs.qza --o-alignment PE_alignment_taxonomy.qza
$ qiime alignment mask --i-alignment PE_alignment_taxonomy.qza --o-masked-alignment PE_masked_alignment.qza
$ qiime phylogeny fasttree --i-alignment PE_masked_alignment.qza --o-tree PE_unrooted_tree.qza
$ qiime phylogeny midpoint-root --i-tree PE_unrooted_tree.qza --o-rooted-tree PE_rooted_tree.qza
Calculate the different beta diversity parameters with the following command:
$ qiime diversity core-metrics-phylogenetic --i-phylogeny PE_rooted_tree.qza --i-table PE-table.qza --p-sampling-depth <90% of the number of reads of the smallest sample in PE-table.qzv> --m-metadata-file metadata.txt --output-dir core_metrics_results
Perform the necessary statistical tests for the beta diversity analyses you require using the following command:

