Protocol for Data Independent Acquisition - Mass spectrometry analysis – a DIA-based Organelle Proteomics
Dario R Alessi, Raja Sekhar Nirujogi, Rotimi Fasimoye, Toan K Phung
Abstract
Purification of intact organelles by previously described methods (dx.doi.org/10.17504/protocols.io.bybjpskn; dx.doi.org/10.17504/protocols.io.6qpvrdjrogmk/v1) allows to profile the organelle proteome using quantitative mass spectrometry. Here we provide a detailed protocol for the Data Independent Acquisition (DIA)-based mass spectrometry (MS) data acquisition method for proteomic profiling of the Golgi. This includes a description of how to construct the nano Liquid chromatography and DIA MS methods as well as a Data Dependent Acquisition (DDA) strategy to generate deep spectral libraries to be able to use in searching the DIA data. In addition, we provide detailed search parameters for database search for both DDA and DIA and downstream MS data analysis.
Attachments
Steps
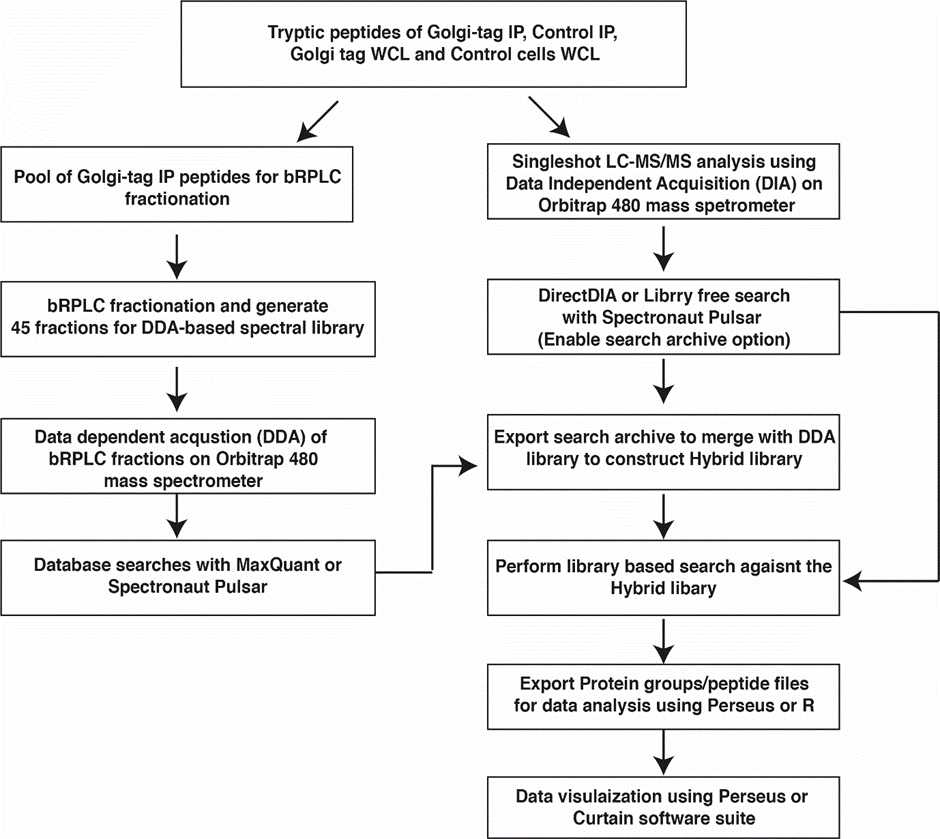
High-pH Reversed-phase Liquid Chromatography fractionation of pooled Golgi-tag IP peptides to generate Spectral library:
Take ~5µg of peptide digest from each of the Golgi-tag IP and Control-IP sample.
Vacuum dry the pooled samples.
Dissolve the peptide digest by adding 120µL of High-pH Solvent-A (10millimolar (mM) Ammonium formate 10.0). Place the sample on a Thermomixer with an agitation at 1800rpm,0h 0m 0s for 0h 30m 0s.
Centrifuge the sample at high speed (17000x g,0h 0m 0s) for 0h 5m 0s at Room temperature.
Take 0.5µL of the sample and verify the pH and transfer the sample into LC-vial.
Ensure the LC-solvent are as Solvent-A (10millimolar (mM) Ammonium formate 10.0); Solvent-B (90% ACN (v/v) in 10millimolar (mM) Ammonium formate 10.0).
Prepare the LC method by following the below gradient:
| A | B | C |
|---|---|---|
| Time (minutes) | Nano pump Flow rate (µl/min) | % Of Solvent-B |
| 0.0 | 0.100 | 3.0 |
| 5.0 | 0.100 | 7.0 |
| 5.5 | 0.100 | 7.0 |
| 10.0 | 0.100 | 10.0 |
| 50.0 | 0.100 | 40.0 |
| 55.0 | 0.100 | 90.0 |
| 62.0 | 0.100 | 90.0 |
| 62.5 | 0.100 | 3.0 |
| 70.0 | 0.100 | 3.0 |
| 70.1 | 0.0100 | 3.0 |
Set the fraction collection time as Start time 0h 5m 5s and End time 1h 2m 0s.
Collect a total of 45 fractions by keeping the fraction collection for 0h 1m 15s for each fraction.
Transfer the fractions into a pre-labelled 1.5mL protein lo binding tubes.
Vacuum dry the samples and freeze in -20 freezer until the LC-MS/MS analysis.
Single shot DIA acquisition on Orbitrap Exploris 480:
Dissolve vacuum dried peptides in 60µL of LC buffer (3% ACN in 0.1% Formic acid) and place the samples on a Thermomixer and mix them at 1800rpm,0h 0m 0s at Room temperature for about 0h 30m 0s.
Take 4µg equivalent of peptide digest and spike 1µL of iRT peptide mix. Adjust the total volume of the sample anywhere between 5µL to 15µL but don’t exceed 15µL. Transfer the sample into glass insert and place them on LC vial.
Construct LC and vDIA MS method as described below using Xaclibur software integrated in Thermo Orbitrap Exploris 480 MS acquisition software suite.
Ensure 2 cm trap column (C18, 5μm, 100A°, 100 µ, 2 cm Nano-viper column # 164564, Thermo Scientific) and 50 cm analytical column (C18, 5micromolar (µM), 50 cm , 100Aº Easy nano spray column # ES903, Thermo Scientific) are equilibrated and verify the column performance by injecting 50ng HeLa or another standard digest.
Nano LC gradient for 2h 25m 0s DIA analysis:
| A | B | C |
|---|---|---|
| Time (minutes) | Nano pump Flow rate (µl/min) | % Of Solvent-B |
| 0.0 | 0.250 | 3.0 |
| 12.0 | 0.250 | 7.0 |
| 115.0 | 0.250 | 25.0 |
| 129.0 | 0.250 | 37.0 |
| 130.0 | 0.250 | 95.0 |
| 135.30 | 0.250 | 95.0 |
| 135.80 | 0.250 | 3.0 |
| 145.0 | 0.250 | 3.0 |
| 145.0 | Stop Run |
Mass spectrometer parameters: Refer below settings to construct variable DIA method:
| A | B | C | D |
|---|---|---|---|
| Method duration | 145 min | ||
| MS Global settings: | |||
| Infusion mode: | Liquid Chromatography | ||
| Expected LC peak width (s): | 20 | ||
| Advanced Peak determination: | TRUE | ||
| Default charge state: | 3 | ||
| Internal mass calibration: | off | Note: If needed enable user defined calibrant ion (Polysilaxolane 445.120025 or enable Easy-IC option | |
| Full scan settings: | |||
| Orbitrap resolution: | 120000 | ||
| Scan range (m/z): | 375-1500 | ||
| RF lens (%): | 40 | ||
| AGC target: | Custom | ||
| Normalized AGC target (%): | 300 | ||
| Maximum injection Time mode: | Custom | ||
| Maximum injection Time (ms): | 30 | ||
| Micro scans: | 1 | ||
| Data type: | Profile | ||
| tMS2 or DIA settings | Isolation offset: | Off | |
| Collision Energy Mode: | Stepped | ||
| Collision Energy Type: | Normalized | ||
| HCD Collision Energy (%): | 25, 28, 32 | ||
| Orbitrap resolution: | 30000 | ||
| Scan range mode: | Define m/z range | ||
| Scan Range (m/z): | 200 - 1200 | Note: Maximum of the matched fragement ions (b series and y-series) fall within this range and if needed this can be modified. | |
| RF Lens (%): | 50 | ||
| AGC target: | Custom | ||
| Normalized AGC target (%): | 3000 | Note: It is recommended to fill the trap with a maximum accumulation of ions (3000% = 3E6 ions) for each of the DIA window to increase the sensitivity | |
| Maximum injection Time mode: | Custom | ||
| Maximum injection Time (ms): | 70 | ||
| Micro scans: | 1 | ||
| Data type: | Profile | ||
| Polarity: | Positive | ||
| Loop control: | N | ||
| N (Number of Spectra): | 24 | We include one full MS1 scan after every 24 DIA scans to accommodate maximum possible MS1 scans | |
| Dynamic RT: | Off | ||
| Time Mode: | Unscheduled |
| A | B | C | D |
|---|---|---|---|
| Scheme of vDIA windows mass list table: | |||
| m/z | z | Isolation Window (mz) | |
| 383.375 | 3 | 66.8 | |
| 423 | 3 | 13.5 | |
| 435 | 3 | 11.5 | |
| 446.5 | 3 | 12.5 | |
| 458 | 3 | 11.5 | |
| 469 | 3 | 11.5 | |
| 480 | 3 | 11.5 | |
| 490.5 | 3 | 10.5 | |
| 501 | 3 | 11.5 | |
| 512 | 3 | 11.5 | |
| 523 | 3 | 11.5 | |
| 533.5 | 3 | 10.5 | |
| 544 | 3 | 11.5 | |
| 554.5 | 3 | 10.5 | |
| 565 | 3 | 11.5 | |
| 575.5 | 3 | 10.5 | |
| 586 | 3 | 11.5 | |
| 597.5 | 3 | 12.5 | |
| 609.5 | 3 | 12.5 | |
| 621.5 | 3 | 12.5 | |
| 633 | 3 | 11.5 | |
| 645 | 3 | 13.5 | |
| 657.5 | 3 | 12.5 | |
| 670.5 | 3 | 14.5 | |
| 684 | 3 | 13.5 | |
| 697 | 3 | 13.5 | |
| 710.5 | 3 | 14.5 | |
| 725.5 | 3 | 16.5 | |
| 741 | 3 | 15.5 | |
| 756.5 | 3 | 16.5 | |
| 773.5 | 3 | 18.5 | |
| 791 | 3 | 17.5 | |
| 808.5 | 3 | 18.5 | |
| 827 | 3 | 19.5 | |
| 846.5 | 3 | 20.5 | |
| 866.5 | 3 | 20.5 | |
| 887.5 | 3 | 22.5 | |
| 910.5 | 3 | 24.5 | |
| 935.5 | 3 | 26.5 | |
| 962.5 | 3 | 28.5 | |
| 992 | 3 | 31.5 | |
| 1025 | 3 | 35.5 | |
| 1063 | 3 | 41.5 | |
| 1108.5 | 3 | 50.5 | |
| 1391.625 | 3 | 516.8 |
Export the MS raw data for database searches by library-free (direct DIA) or library-based as illustrated in the workflow with Biognosys Spectronaut software suite.
Data Dependent Acquisition (DDA) MS analysis to generate Spectral library:
Dissolve vacuum dried peptides of each fraction in 60µL of LC buffer (3% ACN in 0.1% Formic acid) and place the samples on a Thermomixer and mix them at 1800rpm,0h 0m 0s at Room temperature for about 0h 30m 0s.
Take 1µg equivalent of peptide digest and Spike 1µL of iRT peptide mix. Adjust the total volume of the sample anywhere between 5µL to 15µL but don’t exceed 15µL. Transfer the sample into glass insert and place them on LC vial.
Ensure 2 cm trap column (C18, 5μm, 100Ao, 100 µ, 2 cm Nano-viper column # 164564, Thermo Scientific) and 50 cm analytical column (C18, 5micromolar (µM), 50 cm , 100Ao Easy nano spray column # ES903, Thermo Scientific) are equilibrated and verify the column performance by injecting 50ng HeLa or another standard digest.
Nano LC gradient for 1h 25m 0s DDA analysis:
| A | B | C |
|---|---|---|
| Time (minutes) | Nano pump Flow rate (µl/min) | % Of Solvent-B |
| 0.0 | 0.300 | 3.0 |
| 7.0 | 0.300 | 7.0 |
| 60.0 | 0.300 | 22.0 |
| 70.0 | 0.300 | 35.0 |
| 71.0 | 0.300 | 95.0 |
| 78.0 | 0.300 | 95.0 |
| 79.0 | 0.300 | 3.0 |
| 85.0 | 0.300 | 3.0 |
| 85.0 | Stop Run |
Mass spectrometer parameters: Refer below settings to construct DDA method:
| A | B | C |
|---|---|---|
| Method duration | 85 min | |
| MS Global settings: | ||
| Infusion mode: | Liquid Chromatography | |
| Expected LC peak width (s): | 15 | |
| Advanced Peak determination: | TRUE | |
| Default charge state: | 2 | |
| Internal mass calibration: | off | |
| Full scan settings: | ||
| Orbitrap resolution: | 60000 | |
| Scan range (m/z): | 350-1200 | |
| RF lens (%): | 40 | |
| AGC target: | Custom | |
| Normalized AGC target (%): | 300 | |
| Maximum injection Time mode: | Custom | |
| Maximum injection Time (ms): | 28 | |
| Micorscans: | 1 | |
| Data type: | Profile | |
| Polarity: | Positive | |
| Filters: | ||
| MIPS | Monoisotopic peak determination: | Peptide |
| Relax restrictions when too few precursors are found: | FALSE | |
| Intensity | Filter Type: | ntensity Threshold |
| Intensity Threshold: | 1.00E+04 | |
| Charge State | Include charge state(s): | 2 to 6 |
| Include undetermined charge states: | False | |
| Dynamic Exclusion | Dynamic Exclusion Mode: | Custom |
| Exclude after n times: | 1 | |
| Exclusion duration (s): | 45 | |
| Mass Tolerance: | ppm | |
| Low: | 10 | |
| High | 10 | |
| Exclude isotopes: | TRUE | |
| Perform dependent scan on single charge state per precursor only: | FALSE | |
| Data Dependent | Data Dependent Mode: | Cycle Time |
| Time between Master Scans (sec): | 3 | |
| ddMS2 settings | Isolation Window (m/z): | 1.2 |
| Isolation Offset: | Off | |
| Collision Energy Mode: | Fixed | |
| Collision Energy Type: | Normalized | |
| HCD Collision Energy (%): | 30 | |
| Orbitrap resolution: | 15000 | |
| Scan range mode: | Auto | |
| Scan Range (m/z): | 200 - 1200 | |
| AGC target: | Custom | |
| Normalized AGC target (%): | 100 | |
| Maximum injection Time mode: | Custom | |
| Maximum injection Time (ms): | 85 | |
| Micorscans: | 1 | |
| Data type: | Centroid | |
| Polarity: | Positive |
Database searches with MaxQuant for Data Dependent Acquisition (DDA) MS analysis to generate Spectral library:
Export Raw MS data to a Windows server to perform database searches using MaxQuant. Refer the below search parameters for the search.
| A | B |
|---|---|
| Value | |
| Version | 1.6.10.0 |
| Include contaminants | TRUE |
| PSM FDR | 0.01 |
| PSM FDR Crosslink | 0.01 |
| Protein FDR | 0.01 |
| Site FDR | 0.01 |
| Use Normalized Ratios for Occupancy | TRUE |
| Min. peptide Length | 7 |
| Min. score for unmodified peptides | 0 |
| Min. score for modified peptides | 40 |
| Min. delta score for unmodified peptides | 0 |
| Min. delta score for modified peptides | 6 |
| Min. unique peptides | 0 |
| Min. razor peptides | 1 |
| Min. peptides | 1 |
| Use only unmodified peptides and | TRUE |
| Modifications included in protein quantification | Oxidation (M);Acetyl (Protein N-term) |
| Peptides used for protein quantification | Razor |
| Discard unmodified counterpart peptides | TRUE |
| Label min. ratio count | 2 |
| Use delta score | FALSE |
| iBAQ | TRUE |
| iBAQ log fit | TRUE |
| Match between runs | TRUE |
| Matching time window [min] | 0.7 |
| Match ion mobility window [indices] | 0.05 |
| Alignment time window [min] | 20 |
| Alignment ion mobility window [indices] | 1 |
| Find dependent peptides | FALSE |
| Fasta file | D:\Database\20200723-Human-Uniprot.fasta |
| Decoy mode | revert |
| Include contaminants | TRUE |
| Advanced ratios | TRUE |
| Fixed andromeda index folder | |
| Temporary folder | |
| Combined folder location | |
| Second peptides | TRUE |
| Stabilize large LFQ ratios | FALSE |
| Separate LFQ in parameter group | FALSE |
| Require MS/MS for LFQ comparisons | FALSE |
| Calculate peak properties | FALSE |
| Main search max. combinations | 200 |
| Advanced site intensities | TRUE |
| Write msScans table | TRUE |
| Write msmsScans table | TRUE |
| Write ms3Scans table | FALSE |
| Write allPeptides table | TRUE |
| Write mzRange table | TRUE |
| Write pasefMsmsScans table | FALSE |
| Write accumulatedPasefMsmsScans table | FALSE |
| Max. peptide mass [Da] | 4600 |
| Min. peptide length for unspecific search | 8 |
| Max. peptide length for unspecific search | 25 |
| Razor protein FDR | TRUE |
| Disable MD5 | FALSE |
| Max mods in site table | 3 |
| Match unidentified features | FALSE |
| Epsilon score for mutations | |
| Evaluate variant peptides separately | TRUE |
| Variation mode | None |
| MS/MS tol. (FTMS) | 20 ppm |
| Top MS/MS peaks per Da interval. (FTMS) | 12 |
| Da interval. (FTMS) | 100 |
| MS/MS deisotoping (FTMS) | TRUE |
| MS/MS deisotoping tolerance (FTMS) | 7 |
| MS/MS deisotoping tolerance unit (FTMS) | ppm |
| MS/MS higher charges (FTMS) | TRUE |
| MS/MS water loss (FTMS) | TRUE |
| MS/MS ammonia loss (FTMS) | TRUE |
| MS/MS dependent losses (FTMS) | TRUE |
| MS/MS recalibration (FTMS) | TRUE |
| MS/MS tol. (ITMS) | 0.5 Da |
| Top MS/MS peaks per Da interval. (ITMS) | 8 |
| Da interval. (ITMS) | 100 |
| MS/MS deisotoping (ITMS) | FALSE |
| MS/MS deisotoping tolerance (ITMS) | 0.15 |
| MS/MS deisotoping tolerance unit (ITMS) | Da |
| MS/MS higher charges (ITMS) | TRUE |
| MS/MS water loss (ITMS) | TRUE |
| MS/MS ammonia loss (ITMS) | TRUE |
| MS/MS dependent losses (ITMS) | TRUE |
| MS/MS recalibration (ITMS) | FALSE |
| MS/MS tol. (TOF) | 40 ppm |
| Top MS/MS peaks per Da interval. (TOF) | 10 |
| Da interval. (TOF) | 100 |
| MS/MS deisotoping (TOF) | TRUE |
| MS/MS deisotoping tolerance (TOF) | 0.01 |
| MS/MS deisotoping tolerance unit (TOF) | Da |
| MS/MS higher charges (TOF) | TRUE |
| MS/MS water loss (TOF) | TRUE |
| MS/MS ammonia loss (TOF) | TRUE |
| MS/MS dependent losses (TOF) | TRUE |
| MS/MS recalibration (TOF) | FALSE |
| MS/MS tol. (Unknown) | 20 ppm |
| Top MS/MS peaks per Da interval. (Unknown) | 12 |
| Da interval. (Unknown) | 100 |
| MS/MS deisotoping (Unknown) | TRUE |
| MS/MS deisotoping tolerance (Unknown) | 7 |
| MS/MS deisotoping tolerance unit (Unknown) | ppm |
| MS/MS higher charges (Unknown) | TRUE |
| MS/MS water loss (Unknown) | TRUE |
| MS/MS ammonia loss (Unknown) | TRUE |
| MS/MS dependent losses (Unknown) | TRUE |
| MS/MS recalibration (Unknown) | FALSE |
| Site tables | Deamidation (NQ)Sites.txt;Oxidation (M)Sites.txt;Phospho (STY)Sites.txt |
Database searches with Biognosys Spectronaut for Data Dependent Independent Acquisition (DIA) MS analysis (Library free and Library-based search):
Import the msms.txt file form the MaxQuant search output files into Spectronaut to generate a Spectral library.
Alternatively perform a Pulsar search of DDA data to generate a library.
As illustrated in the workflow we recommend doing a direct-DIA or Library free search using Human Uniprot FAST file to construct a hybrid library. Enable search archive option during the direct-DIA search.
Merge the direct-DIA search archive and DDA library to construct a hybrid library and use this library to perform library-based search of the DIA data.
Use the below settings for the library-based DIA search within Spectronaut.
| A | B |
|---|---|
| Spectronaut 15.7.220308.50606 | |
| Computer Name: MRC-DRI-2 | |
| User Domain Name: LIFESCI-AD | |
| User Name: rnirujogi | |
| Analysis Mode: UI | |
| Analysis Type: Peptide-Centric | |
| Settings Used: RN_DIA_Default | |
| Data Extraction | |
| MS1 Mass Tolerance Strategy: | Dynamic |
| Correction Factor: | 1 |
| MS2 Mass Tolerance Strategy: | Dynamic |
| Correction Factor: | 1 |
| Intensity Extraction MS1: | Maximum Intensity |
| Intensity Extraction MS2: | Maximum Intensity |
| XIC Extraction | |
| XIC IM Extraction Window: | Dynamic |
| Correction Factor: | 1 |
| XIC RT Extraction Window: | Dynamic |
| Correction Factor: | 1 |
| Calibration | |
| Calibration Mode: | Automatic |
| MS1 Mass Tolerance Strategy: | System Default |
| MS2 Mass Tolerance Strategy: | System Default |
| Precision iRT: | TRUE |
| iRT <-> RT Regression Type: | Local (Non-Linear) Regression |
| Exclude Deamidated Peptides: | TRUE |
| MZ Extraction Strategy: | Maximum Intensity |
| Allow source specific iRT Calibration: | TRUE |
| Used Biognosys' iRT Kit: | TRUE |
| Calibration Carry-Over: | FALSE |
| Identification | |
| Generate Decoys: | TRUE |
| Decoy Limit Strategy: | Dynamic |
| Library Size Fraction: | 0.1 |
| Decoy Method: | Mutated |
| Preferred Fragment Source: | NN Predicted Fragments |
| Machine Learning: | Per Run |
| Exclude Duplicate Assays: | TRUE |
| Precursor PEP Cutoff: | 0.2 |
| Protein Qvalue Cutoff (Experiment): | 0.01 |
| Protein Qvalue Cutoff (Run): | 0.05 |
| Exclude Single Hit Proteins: | TRUE |
| Pvalue Estimator: | Kernel Density Estimator |
| Precursor Qvalue Cutoff: | 0.01 |
| Single Hit Definition: | By Stripped Sequence |
| Quantification | |
| Interference Correction: | TRUE |
| MS1 Min: | 2 |
| MS2 Min: | 3 |
| Exclude All Multi-Channel Interferences: | TRUE |
| Only Identified Peptides: | TRUE |
| Protein LFQ Method: | Automatic |
| Major (Protein) Grouping: | by Protein Group Id |
| Minor (Peptide) Grouping: | by Stripped Sequence |
| Minor Group Top N: | TRUE |
| Min: | 1 |
| Max: | 3 |
| Minor Group Quantity: | Mean precursor quantity |
| Major Group Top N: | TRUE |
| Min: | 1 |
| Max: | 3 |
| Major Group Quantity: | Mean peptide quantity |
| Quantity MS-Level: | MS2 |
| Quantity Type: | Area |
| Proteotypicity Filter: | None |
| Data Filtering: | Qvalue |
| Cross Run Normalization: | TRUE |
| Row Selection: | Automatic |
| Normalization Strategy: | None |
| Normalization Filter Type: | None |
| PTM Workflow | |
| PTM Localization: | TRUE |
| Probability Cutoff: | 0.75 |
| PTM Analysis: | TRUE |
| Multiplicity: | TRUE |
| Run Clustering: | FALSE |
| PTM Consolidation: | Sum |
| Flanking Region: | 7 |
| Workflow | |
| In-Silico Library Optimization: | FALSE |
| Profiling Strategy: | iRT Profiling |
| Profiling Row Selection: | Minimum Qvalue Row Selection |
| Qvalue Threshold: | 0.01 |
| Profiling Target Selection: | Automatic Selection |
| Carry-over exact Peak Boundaries: | FALSE |
| Unify Peptide Peaks Strategy: | None |
| Multi-Channel Workflow Definition: | From Library Annotation |
| Fallback Option: | Labeled |
| Protein Inference | |
| Protein Inference Workflow: | Automatic |
| Inference Algorithm: | IDPicker |
| Post Analysis | |
| Calculate Sample Correlation Matrix: | TRUE |
| Calculate Explained TIC: | None |
| Gene Ontology: | geneOntology/Ontologies\bgs_default_go basic.obo |
| Differential Abundance Grouping: | Major Group (Quantification Settings) |
| Smallest Quantitative Unit: | Major Group (Quantification Settings) |
| Use All MS-Level Quantities: | FALSE |
| Differential Abundance Testing: | Un-Paired t-test |
| Assume Equa Variance: | FALSE |
| Group-Wise Testing Correction: | FALSE |
| Run Clustering: | TRUE |
| Distance Metric: | Manhattan Distance |
| Linkage Strategy: | Ward's Method |
| Z-score transformation: | FALSE |
| Order Runs by Clustering: | TRUE |
| Pipeline Mode | |
| Post Analysis Reports: | |
| Scoring Histograms: | TRUE |
| Data Completeness Bar Chart: | TRUE |
| Run Identifications Bar Chart: | TRUE |
| CV Density Line Chart: | TRUE |
| CVs Below X Bar Chart: | TRUE |
| Generate SNE File: | TRUE |
| Store Iontraces in SNE: | FALSE |
| Report Schema: | PTMSiteReport (Pivot), RN_PG_Pivot (Pivot), MSStats Report (v 3.7.3)(Normal), Protein Quant (Normal), Protein Quant (Pivot), BGS Factory Report(Normal) |
| Reporting Unit: | Across Experiment |
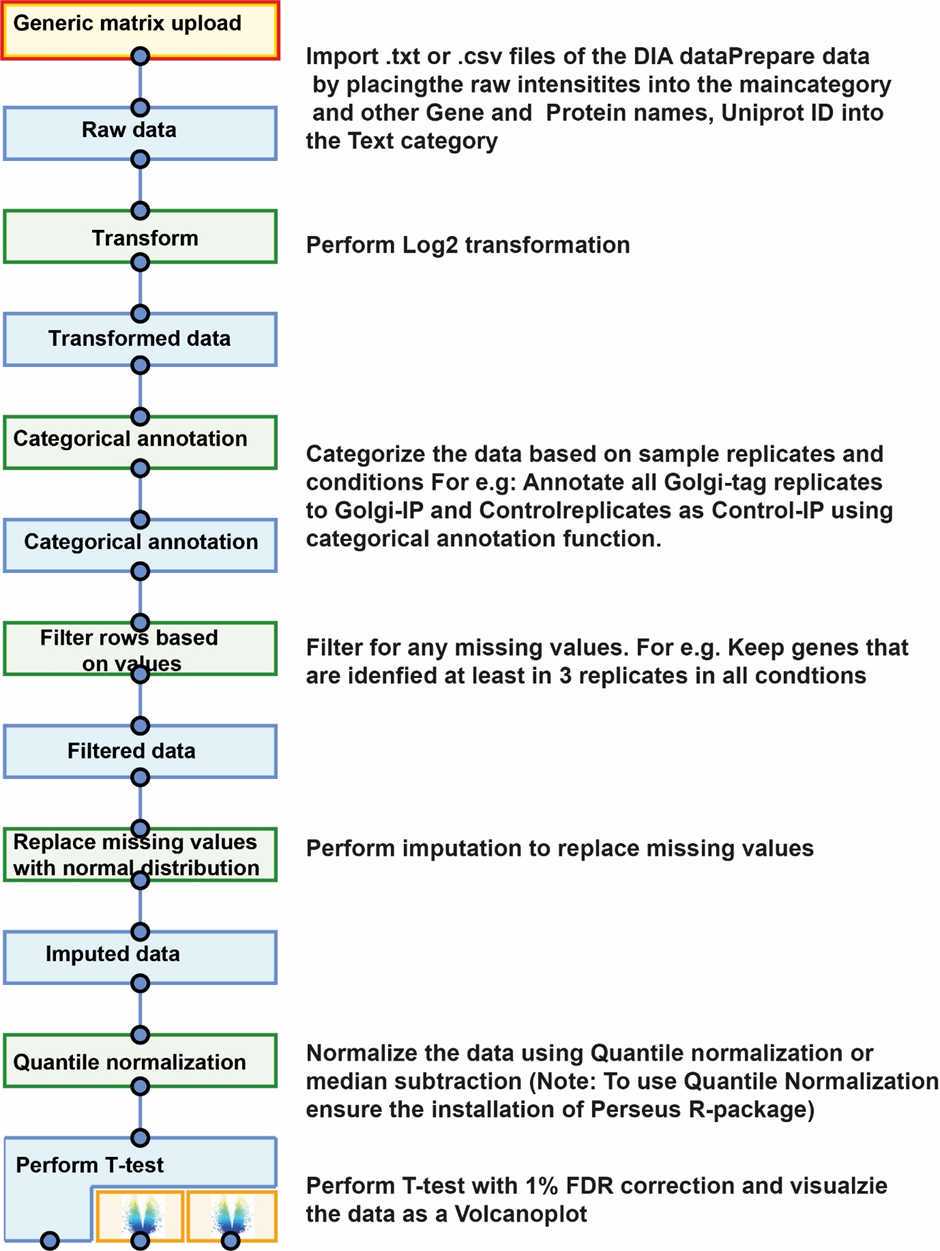
Data analysis of DIA data and data visualization:
Export Protein group tables from Spectronaut in PG Pivot format.
For the Golgi-tag IP data, annotate using a complied list of know Golgi proteins from a resource e.g. (https://compartments.jensenlab.org/Search)) and Uniprot-GO terms.
Prepare the data for differential analysis and this can be done using Perseus software suite (https://maxquant.net/perseus/)..) The basic functionalities of the software and various workflows can be adopted from the published literature (PMID: 27348712) and available tutorials (http://coxdocs.org/doku.php?id=perseus:start)) on Youtube (https://www.youtube.com/c/MaxQuantChannel/featured))
The T-test results can be exported and could be analysed using other software suites such as curtain tool to visualize the volcano plot and associated protein raw intensities for all the conditions, protein domain architecture, STRING interaction prediction and alpha fold prediction.
Optional: In addition to using the Perseus other data quality can be done using custom R or Python Scripts (Provided below) and other relevant packages.
Figure: 1
Scripts - R correlation plot
library(corrplot)
filename <- "//mrc-smb.lifesci.dundee.ac.uk/mrc-group-folder/ALESSI/Toan/For
Golgitag_Paper/For_Pearson_Corr_02.txt"
df <- read.table(filename, header = TRUE, sep="\t")
df <- df[colnames(df)[1:which(colnames(df) == "HA.WCL_06")]]
cor_mat <- cor(as.matrix(df), use="everything")
pdf("//mrc-smb.lifesci.dundee.ac.uk/mrc-group-folder/ALESSI/Toan/For
Golgitag_Paper/For_Pearson_Corr_02.txt.pdf")
corrplot(cor_mat, order="hclust", type="lower", method="ellipse")
dev.off()
Scripts - Python Network Interaction with Cytoscape and Plotly Dash
import dash
import dash_cytoscape as cyto
from dash.dependencies import Input, Output
import dash_core_components as dcc
import dash_html_components as html
import pandas as pd
cyto.load_extra_layouts()
app = dash.Dash(__name__)
server = app.server
def add_individual_protein(df, source, elements):
highest = df["Difference"].max()
n = 0
for i, r in df.iterrows():
if n < 15:
opacity = r["Difference"]/highest
elements.append({'data': {'id': r["Gene.names"], 'label': r["Gene.names"], 'color': f"rgba(136, 86, 167,{opacity})", "opacity": opacity}, 'classes': 'protein'})
elements.append(
{'data': {'source': source, 'target': r["Gene.names"], 'color': f"rgba(136, 86, 167,{opacity})", "opacity": opacity}, 'classes': 'protein-edge'},)
else:
break
n += 1
def add_groups_enriched(edf, elements):
edf = edf.sort_values(by="Difference", ascending=False)
golgi = edf[(edf["Golgi"] == "+")]
#golgi = edf[(edf["C: Golgi"] == "+")]
golgi_count = len(golgi.index)
print(golgi_count)
glyco = golgi[golgi["Glycosylation"] == "+"]
#glyco = golgi[golgi["Glycosylation genes"] == "+"]
glyco_count = len(glyco.index)
print(glyco_count)
phospha = golgi[golgi["Phosphatases"] == "+"]
phospha_count = len(phospha.index)
kinases = golgi[(golgi["Kinases"] == "+") | (golgi["Dark.kinase"] == "+")]
#kinases = golgi[(golgi["Kinases"] == "+") | (golgi["Dark Kinases"] == "+")]
kinases_count = len(kinases.index)
ubi = golgi[golgi["Ub.Pathway"] == "+"]
ubi_count = len(ubi.index)
l = [
{'data': {'id': 'enriched-golgi', 'label': f'Golgi: {golgi_count}', "size": golgi_count * block},
'classes': 'golgi enriched'},
#{'data': {'source': 'significant', 'target': 'enriched-golgi'}, 'classes': 'significant-edge'},
{'data': {'id': 'enriched-glyco', 'label': f'Glycosylation genes: {glyco_count}',
"size": glyco_count * block}, 'classes': 'golgi enriched'},
{'data': {'id': 'enriched-phospha', 'label': f'Phosphatases: {phospha_count}',
"size": phospha_count * block}, 'classes': 'golgi enriched'},
{'data': {'id': 'enriched-kinase', 'label': f'Kinases: {kinases_count}', "size": kinases_count * block},
'classes': 'golgi enriched'},
{'data': {'id': 'ubi', 'label': f'Ubiquitin components: {ubi_count}', "size": ubi_count * block},
'classes': 'golgi enriched'},
{'data': {'source': 'enriched-golgi', 'target': 'enriched-glyco'}, 'classes': 'golgi-edge enriched'},
{'data': {'source': 'enriched-golgi', 'target': 'enriched-phospha'},
'classes': 'golgi-edge enriched'},
{'data': {'source': 'enriched-golgi', 'target': 'enriched-kinase'},
'classes': 'golgi-edge enriched'},
{'data': {'source': 'enriched-golgi', 'target': 'ubi'},
'classes': 'golgi-edge enriched'},
]
for i in l:
elements.append(i)
add_individual_protein(glyco, "enriched-glyco", elements)
add_individual_protein(phospha, "enriched-phospha", elements)
add_individual_protein(kinases, "enriched-kinase", elements)
add_individual_protein(ubi, "ubi", elements)
def add_groups_not_enriched(edf, elements):
edf = edf.sort_values(by="Difference", ascending=False)
golgi = edf[(edf["Golgi"] != "+")]
#golgi = edf[(edf["C: Golgi"] != "+")]
golgi_count = len(golgi.index)
print(golgi_count)
glyco = golgi[golgi["Glycosylation"] == "+"]
#glyco = golgi[golgi["Glycosylation genes"] == "+"]
glyco_count = len(glyco.index)
print(glyco_count)
phospha = golgi[golgi["Phosphatases"] == "+"]
phospha_count = len(phospha.index)
kinases = golgi[(golgi["Kinases"] == "+") | (golgi["Dark.kinase"] == "+")]
#kinases = golgi[(golgi["Kinases"] == "+") | (golgi["Dark Kinases"] == "+")]
kinases_count = len(kinases.index)
ubi = golgi[golgi["Ub.Pathway"] == "+"]
ubi_count = len(ubi.index)
l = [
{'data': {'id': 'non-enriched-golgi', 'label': f'Non-golgi: {golgi_count}', "size": golgi_count * block}, 'classes': 'not-golgi not-enriched'},
#{'data': {'source': 'significant', 'target': 'non-enriched-golgi'}, 'classes': 'not-golgi significant-edge'},
{'data': {'id': 'non-enriched-glyco', 'label': f'Glycosylation genes: {glyco_count}',
"size": glyco_count * block}, 'classes': 'not-golgi not-enriched'},
{'data': {'id': 'non-enriched-phospha', 'label': f'Phosphatases: {phospha_count}',
"size": phospha_count * block}, 'classes': 'not-golgi not-enriched'},
{'data': {'id': 'non-enriched-kinase', 'label': f'Kinases: {kinases_count}', "size": kinases_count * block},
'classes': 'not-golgi not-enriched'},
{'data': {'id': 'non-enriched-ubi', 'label': f'Ubiquitin components: {ubi_count}', "size": ubi_count * block},
'classes': 'not-golgi not-enriched'},
{'data': {'source': 'non-enriched-golgi', 'target': 'non-enriched-glyco'}, 'classes': 'not-golgi-edge not-enriched'},
{'data': {'source': 'non-enriched-golgi', 'target': 'non-enriched-phospha'}, 'classes': 'not-golgi-edge not-enriched'},
{'data': {'source': 'non-enriched-golgi', 'target': 'non-enriched-kinase'}, 'classes': 'not-golgi-edge not-enriched'},
{'data': {'source': 'non-enriched-golgi', 'target': 'non-enriched-ubi'},
'classes': 'not-golgi-edge not-enriched'},
]
for i in l:
elements.append(i)
add_individual_protein(glyco, "non-enriched-glyco", elements)
add_individual_protein(phospha, "non-enriched-phospha", elements)
add_individual_protein(kinases, "non-enriched-kinase", elements)
add_individual_protein(ubi, "non-enriched-ubi", elements)
block = 0.2
#df = pd.read_csv(r"C:\Users\toanp\Downloads\All enriched_For Network.txt", sep="\t")
#df = pd.read_csv(r"C:\Users\toanp\Downloads\GT-IP_Mock-IP_tTest.txt", sep="\t")
df = pd.read_csv(r"C:\Users\toanp\Downloads\GT-IP_WCL_tTest.txt", sep="\t")
df = df[(df["Significant"]=="+")&(df["Difference"] >= 1)]
elements = [
#{'data': {'id': 'significant', 'label': f'Significant: {len(df.index)}', "size": len(df.index) * block}, 'classes': 'significant'},
]
add_groups_enriched(df, elements)
add_groups_not_enriched(df, elements)
app.layout = html.Div([
cyto.Cytoscape(
id='cytoscape',
elements=elements,
layout={'name': 'cose', 'idealEdgeLength': 20},
style={'width': '2000px', 'height': '2000px'},
stylesheet=[
{
'selector': '.significant',
'style': {
'shape': 'ellipse',
'background-color': 'rgb(173, 218, 226)',
}
},
{
'selector': '.not-golgi',
'style': {
'shape': 'ellipse',
'background-color': 'rgb(255, 154, 162)',
}
},
{
'selector': '.not-golgi-edge',
'style': {
'curve-style': 'straight-triangle',
"width": 5,
'line-color': 'rgb(255, 154, 162)',
}
},
{
'selector': '.golgi',
'style': {
'shape': 'ellipse',
'background-color': 'rgb(255, 218, 193)',
}
},
{
'selector': '.golgi-edge',
'style': {
'curve-style': 'straight-triangle',
"width": 5,
'line-color': 'rgb(255, 218, 193)',
}
},
{
'selector': '.protein',
'style': {
'shape': 'ellipse',
'background-color': 'data(color)',
'background-opacity': 'data(opacity)',
'line-color': 'black'
}
},
{
'selector': '.protein-edge',
'style': {
'line-color': 'data(color)',
'opacity': 'data(opacity)',
}
},
{
'selector': 'node',
'style': {
"content": "data(label)",
"width": "data(size)",
"height": "data(size)",
}
},
{
'selector': '.enriched',
'style': {
'shape': 'ellipse',
'background-color': 'rgb(255, 154, 162)',
'line-color': 'rgb(255, 154, 162)',
}
},
{
'selector': '.not-enriched',
'style': {
'shape': 'ellipse',
'background-color': 'rgb(255, 218, 193)',
'line-color': 'rgb(255, 218, 193)',
}
}
]
),
html.Div([html.Button("as svg", id="btn-get-svg")])
])
print(elements)
@app.callback(
Output('image-text', 'children'),
Input('cytoscape', 'imageData'),
)
def put_image_string(data):
return data
@app.callback(
Output("cytoscape", "generateImage"),
[
Input("btn-get-svg", "n_clicks"),
])
def get_image(get_svg_clicks):
# File type to output of 'svg, 'png', 'jpg', or 'jpeg' (alias of 'jpg')
# 'store': Stores the image data in 'imageDataf' !only jpg/png are supported
# 'download'`: Downloads the image as a file with all data handling
# 'both'`: Stores image data and downloads image as file.
ctx = dash.callback_context
if ctx.triggered:
input_id = ctx.triggered[0]["prop_id"].split(".")[0]
if input_id != "tabs":
action = "download"
ftype = input_id.split("-")[-1]
return {
'type': 'svg',
'action': 'download'
}
return {
'type': 'png',
'action': 'store'
}
if __name__ == "__main__":
app.run_server(debug=True)