Protocol for DNA Extraction from Tepary Bean
Magdalena M Julkowska, Aparna Srinivasan
Abstract
Protocol to extract high quality DNA from Tepary Bean leaves using modified CTAB method.
Steps
Collection of leaf sample
Collect 100 mg of young leaf from Tepary Bean plants in a 2ml eppendorf tubes containing two sterile glass beads and freeze immediately in liquid nitrogen.
DNA Extraction
After 30 min, remove the tubes from water bath and centrifuge at 14000 rpm for 15 min.
Transfer cleared lysate of 500 μl to a new 1.5 ml tube. Add RNase (10 mg/ml) and keep at 37 °C for 30 min.
Add equal volume of Phenol: Chloroform (phenol:chloroform kit), and vortex well.

Centrifuge at 14000 rpm for 15 min. Transfer the upper aqueous layer to a new 1.5 ml Ep tube.

Do not touch below layers while transferring the aqueous phase.
Repeat the Phenol: Chloroform alcohol step if the aqueous phase is not clear.
Add 300 μl of 3M Sodium Acetate and 500 μl 70 % ethanol and centrifuge at 14000 rpm for 5 min.
Discard the supernatant.
Add 200 μl 70 % Ethanol and centrifuge at 14000 rpm for 5 min.
Discard the supernatant.
Air-dry the pellet, dissolve the pellet in 50 μl TE Buffer.
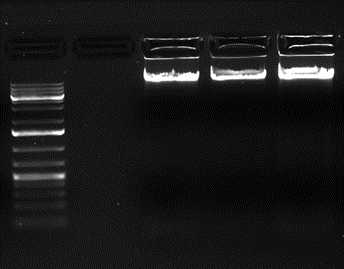
DNA is run on a 0.8% Agarose gel to check whether it is degraded or having RNA contamination.
If RNA is present, treat the samples with RNAse A and precipitate again from phenol:chloroform step 4.
Nucleic acid concentration is measured in Nanodrop, and Ratio of A260/A230 with 1.8-2.0 indicates purity of DNA.
In case the values are <1.8, it indicates contamination such as carbohydrate or phenol.
Store DNA sample at -80 °C until use.