Processing frozen archival human DNA samples for large-scale SQK-LSK114 Oxford Nanopore long-read DNA sequencing SOP v1
Kimberly Paquette, Kimberley J Billingsley, Laksh Malik, on behalf of the CARD Long-read Team, Breeana Baker, Alicia Wenghöfer, Cedric Kouam
Long-read sequencing
Oxford Nanopore sequencing
High molecular weight DNA extraction
DNA extraction
Human blood extraction
Whole blood extraction
DNA size selection
DNA shearing
Disclaimer
In development
We are still developing and optimizing this protocol.
Abstract
Abstract:
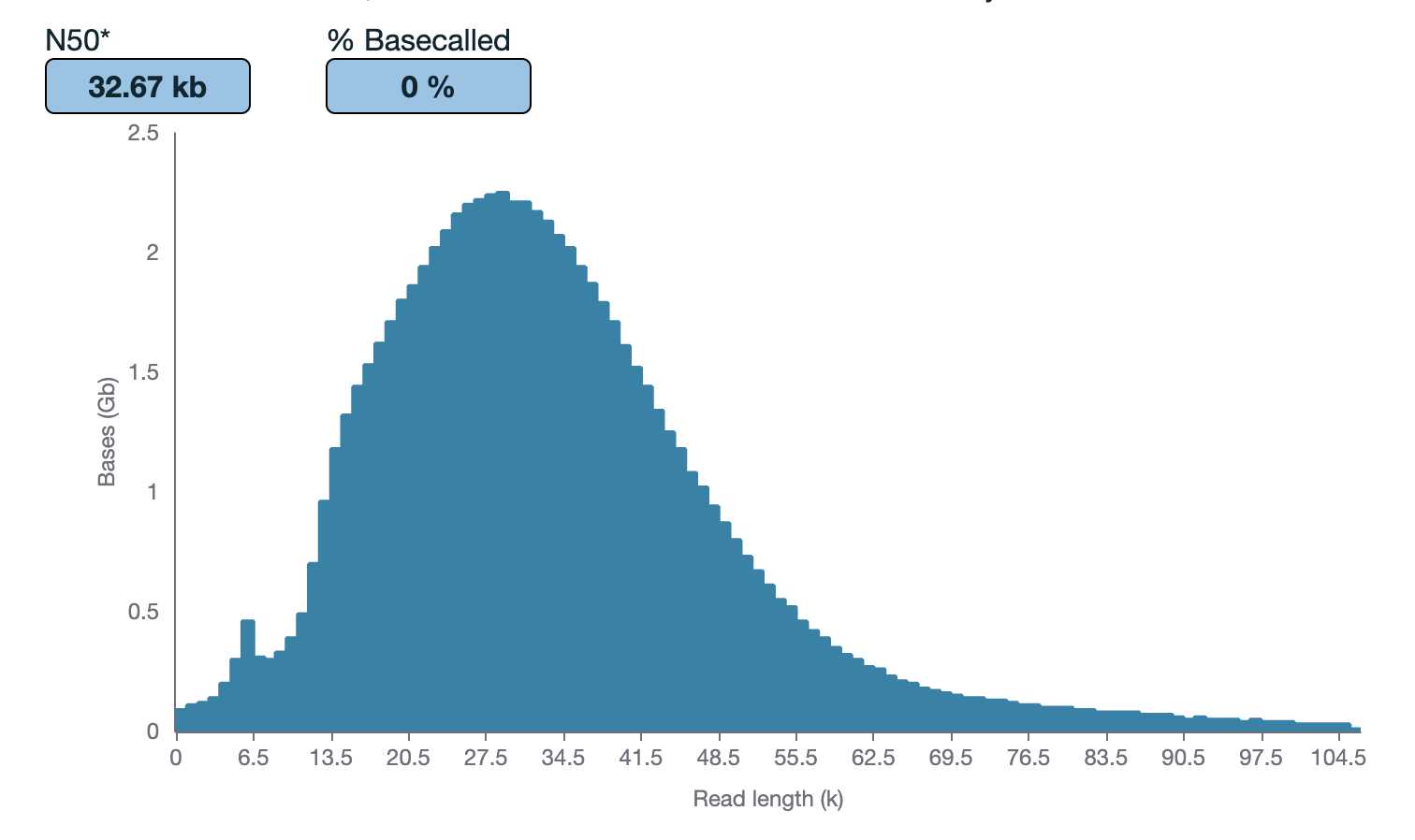
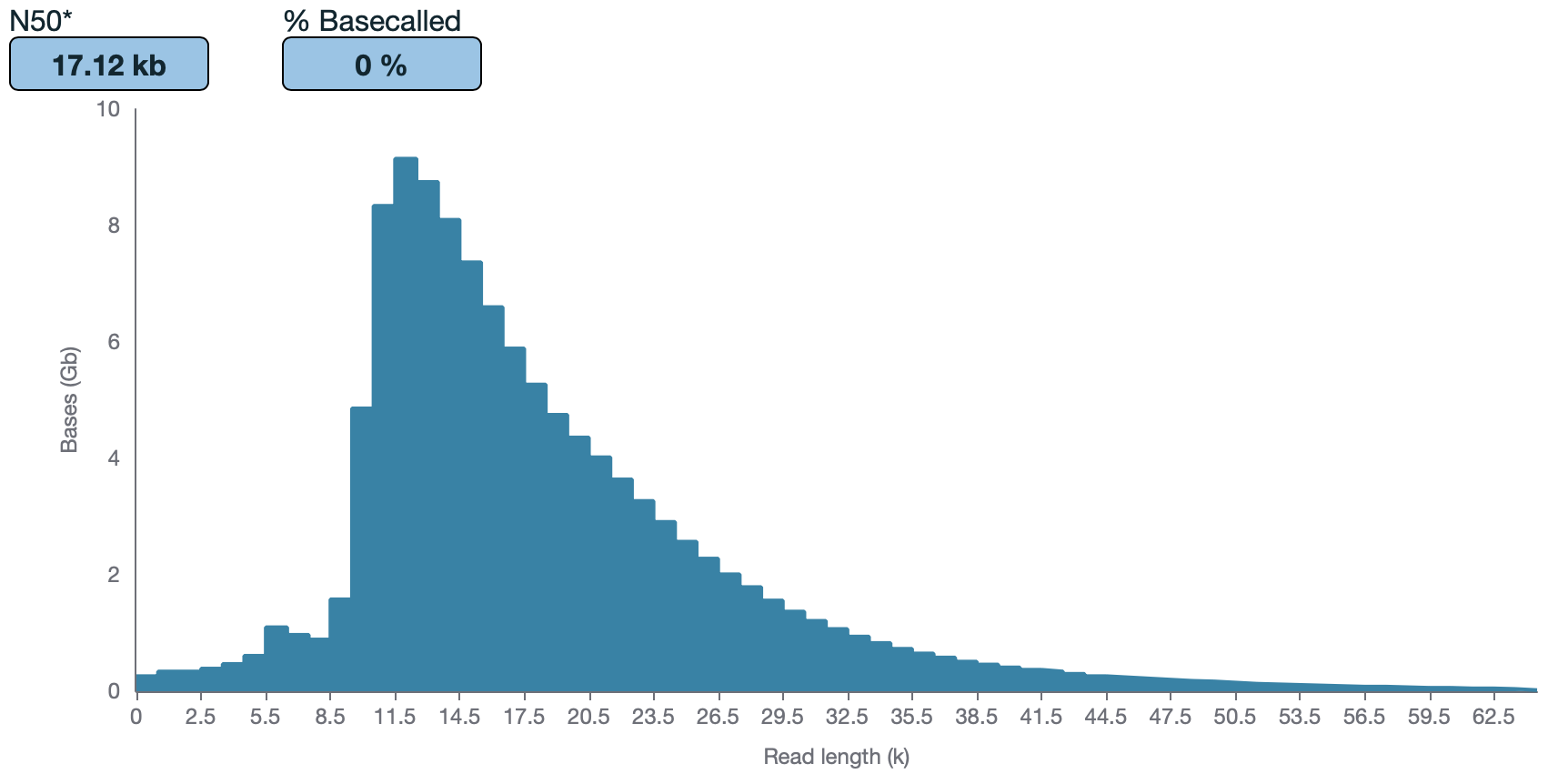
As part of the GP2 monogenic network we will generate long-read sequencing data to better understand the genetic architecture of Parkinson's disease. To generate this large-scale Nanopore data we have developed a protocol for processing and long-read sequencing frozen human DNA samples, ideally targeting an N50 of ~30kb and ~30X coverage. However, with archival human DNA samples, we usually see a drop in DNA quality in terms of DNA length. Therefore, this protocol is focused on attempting to achieve the highest N50 and coverage possible from the input available.

Steps
Part 1: Thaw DNA samples On ice (~ ) 0h 5m 0s)
Note: Ideally for best results, DNA needs to be extracted with high-molecular weight methods straight from tissue. See dx.doi.org/10.17504/protocols.io.5qpvo3639v4o/v1 or dx.doi.org/10.17504/protocols.io.6qpvr347bvmk/v1 dx.doi.org/10.17504/protocols.io.6qpvr347bvmk/v1 or dx.doi.org/10.17504/protocols.io.kxygx3zzog8j/v1 dx.doi.org/10.17504/protocols.io.kxygx3zzog8j/v1 based on tissue type. However, if only archival DNA samples are available, continue with this protocol.
Remove the old DNA samples stored at -80°C and thaw them On ice.
Part 2: DNA Quantification (measuring DNA concentration and size) (~ for 16 samples) 0h 30m 0sfor 16 samples)
Note: This step is to determine if shearing and/or size selection are needed. This is based off fragment length distribution and DNA mass available per sample.
Note: This duration does not include the time it takes to size the samples. The total time it takes to size 16 samples on the TapeStation or the Femto Pulse is ~0h 45m 0s or ~3h 30m 0s, respectively.
Pipette mix samples 10x.
Quantify by taking a single measurement on the Qubit Flex Fluorometer . Use 1µL or 2µL of the sample out of the middle of the pipette-mixed aliquot.
Quantify the purity of the samples (absorbance ratios (260/280 and 260/230)) by taking a single measurement on the NanoDrop . Use 1µLof the sample out of the middle of the pipette-mixed aliquot.
Size on the Agilent Tapestation 4200 or Agilent Femto Pulse System . The expected size range for samples post-extraction is >40kb .
Note for the use of Agilent Femto Pulse System: use DNA sample dilutions between 0.005-0.5ng/ul in 1x TE or EB-Buffer.
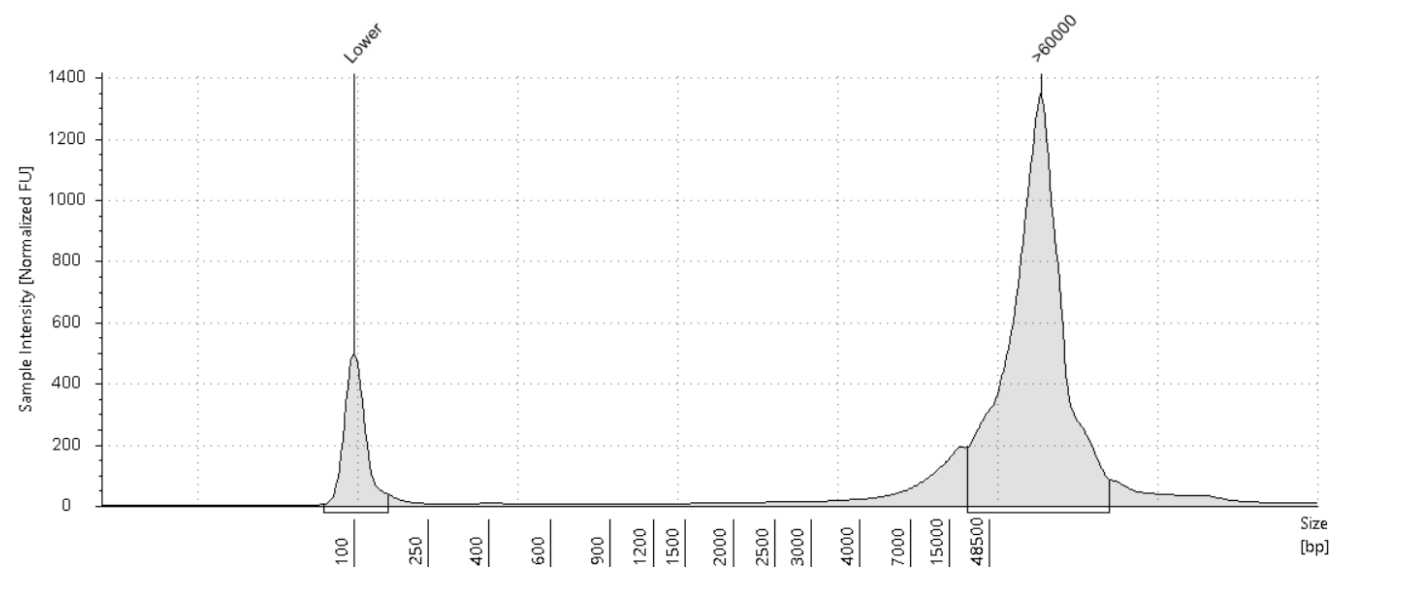
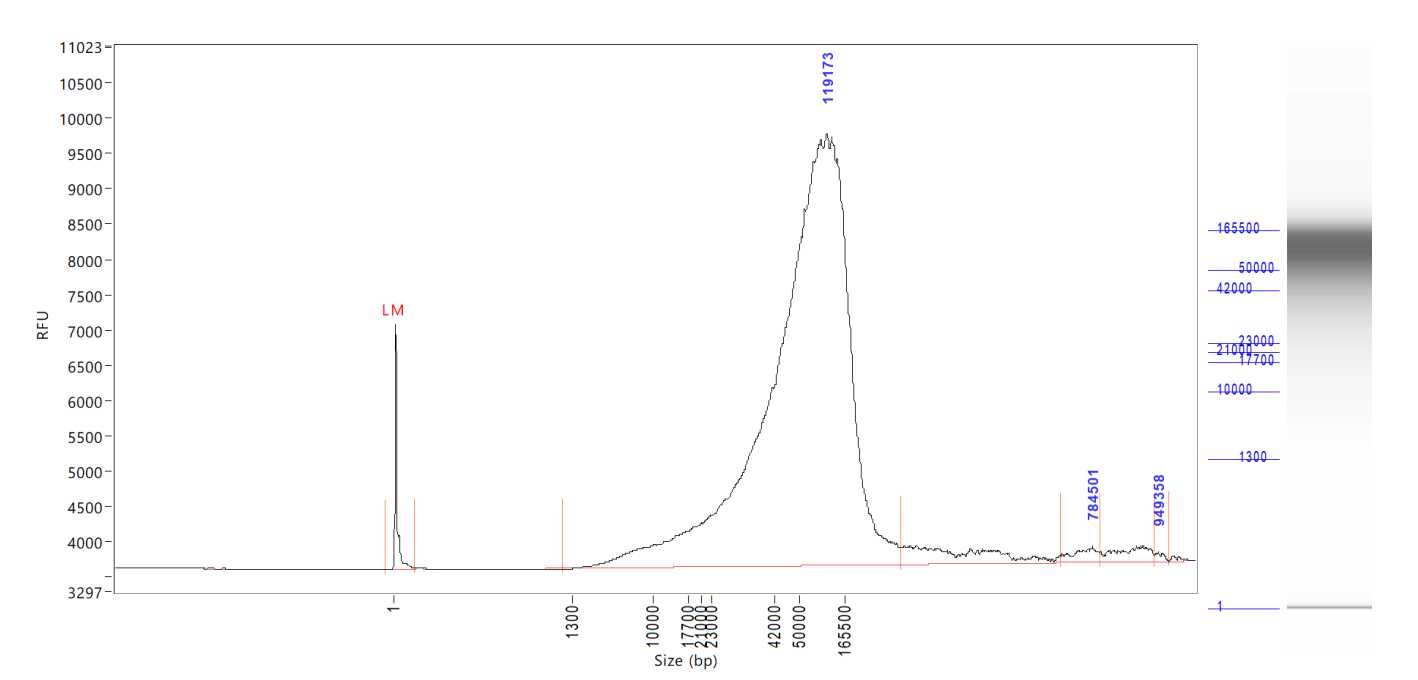
Note: It is recommended to use the Femto Pulse System to get a better representation of the size distribution of the DNA samples.
Once all the samples are properly quantified, move on to size selection.
Part 3: Shearing (~ for 8 samples) 3h 0m 0sfor 8 samples)
We will be using the Diagenode Megaruptor 3 Shearing Kit (E07010003).
In a Megaruptor 3 shearing tube, make up the size selected sample to 100-150µL and 100-150ng/ulwith TE Buffer or water. Do not go over maximum volume of 180µL and maximum concentration of 150ng/ul.
Note: It is ok if target concentration cannot be reached for all samples. If concentration differs from this range, make sure the Megaruptor 3 shearing settings are updated to reflect your sample concentration and volume. If shearing more than one sample try to get all the concentrations to be as close as possible to this target range and/or to each other. Use the Megaruptor 3 for the different samples in the same range of concentration and volume:
- range of the concentration: +/-
10ng/ul - range of volume: +/-
15µL
Note: We recommend taking in a larger concentration into shearing since you will lose some sample when removing the syringe post-shear. If the sample concentration is higher than 70ng/ul diluting it with TE buffer or water to 150µL would make it less than 70ng/ul, it is suggested to dilute to 100µL rather than 150µL. If the sample concentration does not reach 70ng/ul, it is suggested to take all sample into shearing and do not dilute with TE buffer or water. Make sure Megaruptor 3 shearing settings reflect your exact sample volume.
Note: If the sample size is >30kb shear it 1x. If the sample size >100kb shear it 2x. If the sample size is <30kb the shearing step can be skipped. If DNA is viscous and high molecular weight (>100kb ), use fluid + needles (gray). Otherwise the standard needles (black) can be used.
Attach the Megaruptor 3 shearing syringe onto the tube and push the entire item into the MR3 slots until it fits snugly. If running fewer than 8 samples, put the tubes in the 1st and/or 8th slots, working your way in.
Note: Samples should always be balanced. If running an odd number of samples, samples can be balanced with an empty corresponding tube and syringe.
Shear once or twice at speed 20 (takes ~3h 0m 0s if the MR3 is set to 150µL volume and sheared twice; takes 1h 0m 0s if the MR3 is set to 100µL volume and sheared once).
Note : There is no setting for Megaruptor to shear twice. It requires the cycle to be manually restarted for shearing a second time.
Once finished, remove the sample from the Megaruptor 3 and carefully remove the syringe from the tube. Make sure the plunger is fully depressed in order to avoid losing sample. Use a P20 pipette to aspirate any leftover sample on the syringe.
Avoid any vortexing of the DNA from this point on to avoid any unnecessary further shearing. Instead mix by gently flicking the tube and spin down.
Part 4: Post-Shear DNA Quantification (~ for 8 samples) 0h 10m 0sfor 8 samples)
Quantify the samples on the Qubit and size using the Agilent TapeStation 4200 or the Agilent Femto Pulse System . Although the targeted size is 30kb, the observed size range for samples post-shear is between 30-70kb. The samples usually still sequence around 30kb .
Part 5: Size selecting using Blue Pippin (~ 4h 0m 0s for 4 samples)
Using TE, dilute up to 10µgof sheared DNA sample into a final volume of 60µL.
Note: The standard protocol to start with is 60/20 (60µL + 20µL standard loading solution). The sample dilution can also be made up to 30µL for the 30/10 standard loading protocol or 70µL for the 70/10 (use 2x loading solution) loading protocol. See Table 4 for more information.
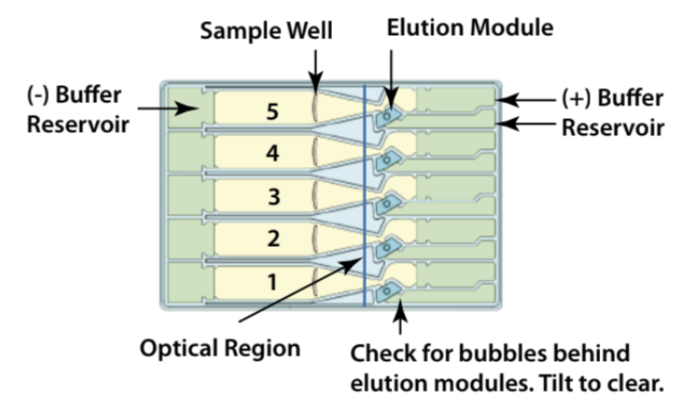
Place the calibration fixture onto the optical nest. The dark side of the fixture must be down, and completely cover all the LED detectors.
Close the lid and press "Calibrate". The calibration status will read "Calibration Underway" and the LED photocurrents will automatically adjust to a factory setting. Once calibration is successful the "Calibration Status" field will contain the message "Calibration OK". In the resulting sub-window, check that the calibration target box ("Target I ph,mA") at the top of the window reads "0.60". If not, manually change the target value to "0.60". Press "EXIT".
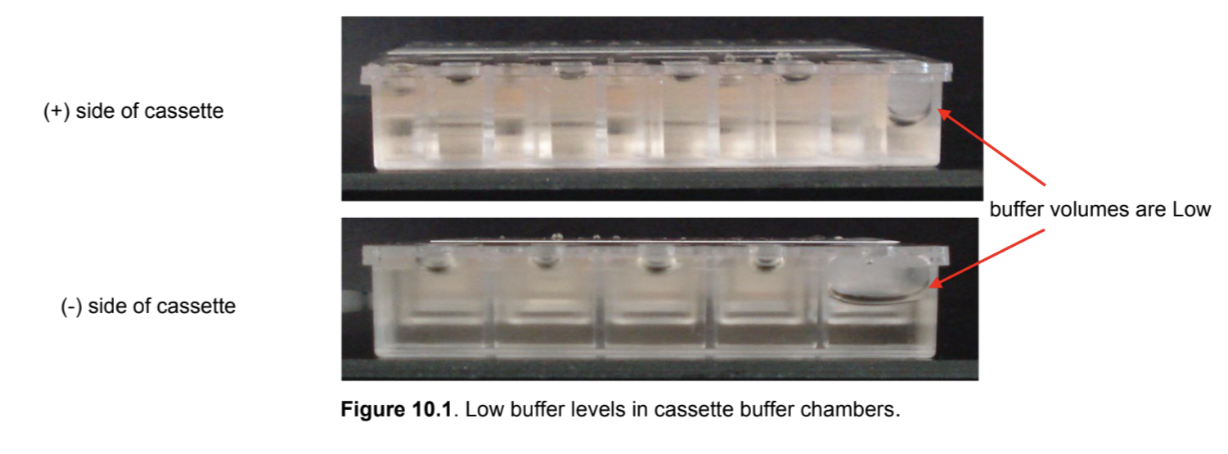
Remove the cassette from the packaging. Inspect the buffer chambers to make sure there are no chambers that are too low. If so, make a mental note of it and add more buffer when you load the cassette in step 5.16.
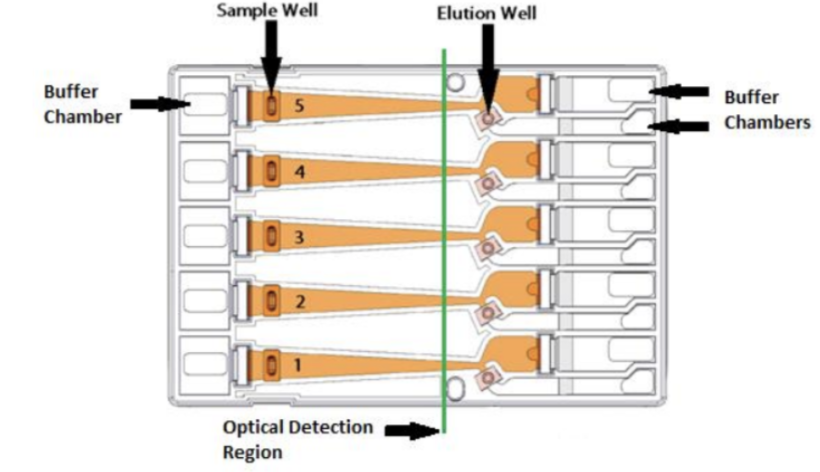
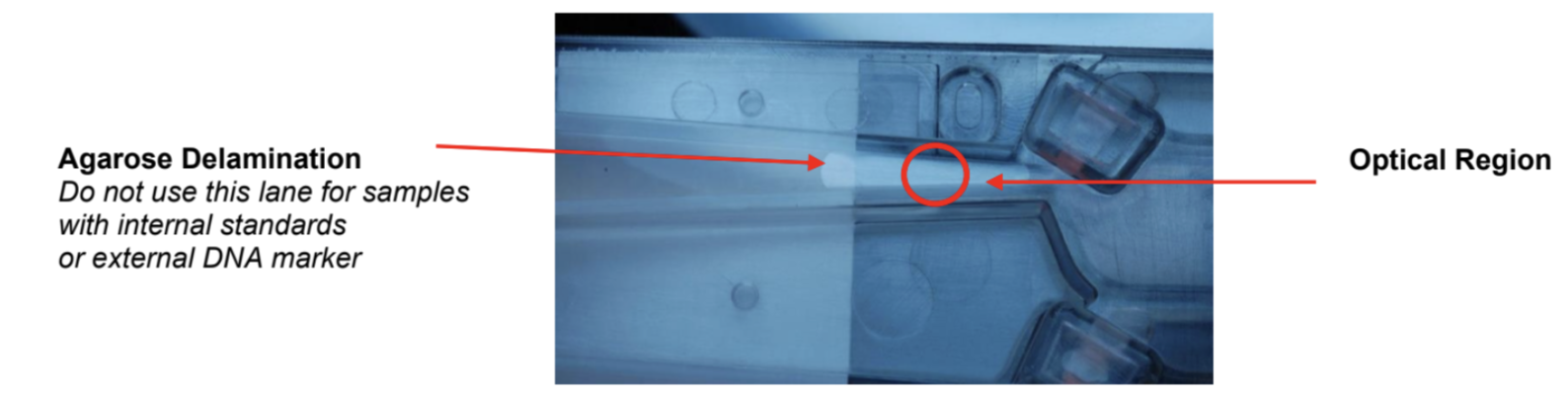
Place the cassette under a light source and inspect for bubbles due to delimitation of agarose from the bottom of the cassette in the region used for optical detection of DNA. If a bubble is detected, do not use the affected lane to run the DNA reference marker. Affected lanes can be used for size selection of sample DNA.
If there are any trapped air bubbles behind the elution wells, tilt the cassette, sample well side down, and gently tap the sides to release the bubbles to the top in direction to the Buffer chambers.
Remove 450µL of buffer from the large buffer reservoirs to reduce the amount of leakage after the run; however, if any of the left or right buffer reservoirs appear to be extremely low from step 5.12, do not remove buffer. Instead, add extra buffer to match to the level of other ones.
Remove both buffer chamber adhesive strips by using the white tabs and pulling the strips slowly towards the front of the instrument until they are removed. To prevent any issues, such as contamination, it may be a good idea to leave the adhesive strips on the sample wells until you are ready to use them. Wipe excess liquid from around the elution modules (not in the well).
Completely remove all buffer from each elution module (around 90µL). Move the pipette tip slowly as you aspirate liquid from the corners of the elution modules to avoid piercing them. You can use the same pipette tip to remove the buffer from each elution module but remember to go back with a new pipette tip to remove any leftovers and reintroduced bubbles. You can also change tips between wells.
At this time, remove the loading solution from the 4°C to allow it to reach Room temperature.
In the Blue Pippin software, click the "Protocol Editor" Tab and click the folder icon to add a new protocol.
Add80µL of the provided electrophoresis buffer to each well. Change tips between wells. To avoid trapping more air bubbles, it is good to go slowly raise the pipette tip as the liquid is being dispensed.
Seal the elution modules with the provided adhesive tape strip and seal. Use a edge to tightly seal and remove any wrinkles.
Close the lid and person Continuity Test by clicking the "Test" button in the "Main" tab.The test sub-window will open and the current in each channel will automatically be measured. If the test returns a "PASS" message, click "RETURN" to exit. if there is a "FAILED" message, make sure there is buffer in every chamber and elution module.
Remove adhesive strips of sample wells by using the white tabs and pulling the strips slowly towards the front of the instrument until they are removed.
Remove 40µL of buffer from the marker well. Remove 80µL (if sample volume is =60µL + 20µL LS) from the sample wells (see Table 4 for additional protocols). Take care not to pierce the agarose with the pipette tip. When removing buffer, it may be good to immerse the tip just below the buffer surface and follow the liquid down with the tip as the buffer is being removed.
At this time, remove the DNA Marker from the 4°C . Do not leave out for any longer than 0h 5m 0s. If you will not be able to use it by them, remove the DNA marker from the 4°C at a later step.
Add 40µL of marker to the marker well and return the marker to the 4°C.
Add loading solution (LS) to the sample tube based on the sample volume, pipette mix, and pulse-centrifuge:
| A | B | C | D |
|---|---|---|---|
| 30 uL | Standard | 10 uL | 40 uL |
| 60 uL | Standard | 20 uL | 80 uL |
| 70 uL | 2x | 10 uL | 80 uL |
Table 4. Sample volume and loading solution (LS) volume.
Note: For the 70/10uL sample to LS mix, the 2x LS is needed.
When adding samples to the respective wells, place the tip of the pipette just below the surface of the buffer and slowly dispense. Do not be concerned if the sample well slightly overfills. The density of the sample will allow it to sink during the run. Sample wells should be completely filled to the top with buffer. If any wells are under-filled, top them off with additional electrophoresis buffer.
Close the lid and press "Start" on the "Main" tab. A checklist screen will appear reminding the user to do the LED calibration, which should have been done earlier. Press "Start".
For best recovery, wait at least 0h 45m 0s (better overnight) after the end of the run before removing samples from the elution modules. Slowly, remove samples. The volumes should be close to 80µL
Click "Casette Type" folder icon and select the greyed out option 0.75 Agarose Dye-free. Then choose "10 Kb High Pass Plus Marker U1" or "15 Kb High Pass Plus Marker U1" based on the desired protocol.
Additional 10-30% yield can often be achieved by adding 80µL of the provided Tween solution to each empty elution module. Wait 0h 1m 0s, then remove the solution and place in separate tubes.
Preventative Maintenance - Electrodes should be rinsed after 5 runs or once per week. Fill the provided rinse cassette with deionized water. Load the rinse cassette onto the sample tray and close the lid. Allow the electrodes to rinse for several seconds. Open the lid and remove the cassette.
Enter the Sample ID into the "Sample ID" fields. Make sure that the check box for "End Run when Elution is Completed" is selected. Enter Marker for the Marker Well.
Note: Lane 4 is the preferred lane for the Marker.
In the "Range" column, make sure that all of the rectangle blocks are selected (light grey color) except of the Marker well (dark grey).
Note : The Marker lane requires a manual deselection of the row (changes color of row to dark grey).
In the Reference Lane field, choose the Marker well number (lane 4) and press the "APPLY REFERENCE TO ALL LANES" button.
Enter the number "10000" (10 kb = 10000) or "15000" (15 kb = 15000) or in the BP "Start" field. The BP "End" and "Target" values are cassettes will still collect anything past the start field.
Click "Save As" to name and save the run.
In the software, click the "Main" Tab.
Part 6: Post-BluePippin DNA Quantification (~ for 8 samples) 0h 10m 0s for 8 samples)
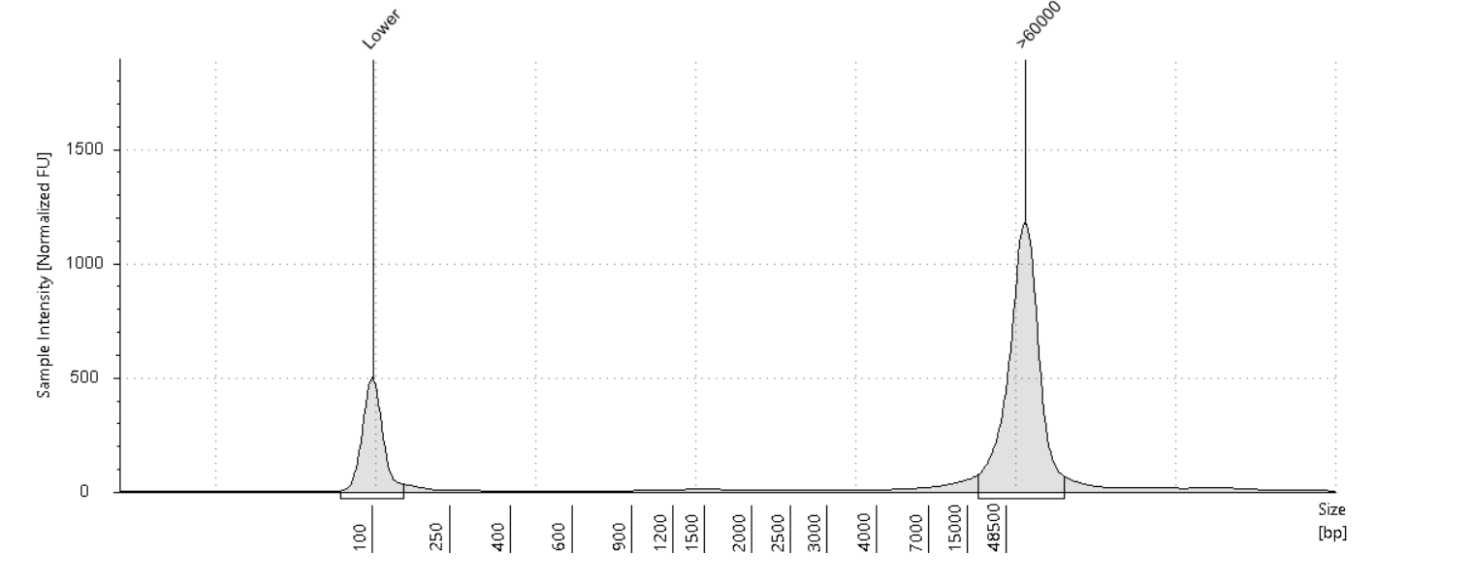
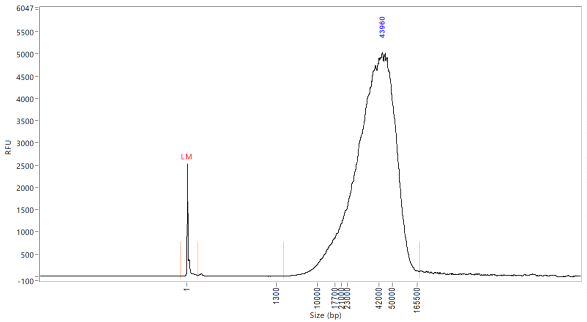
Quantify the samples on the Qubit and NanoDrop and size using the Agilent TapeStation 4200 or the Agilent Femto Pulse System. Although the targeted size is 30kb, the observed size range for samples post-shear is between 30-70kb. The samples usually still sequence around 30kb.


At this point, ideally 2.5µg of DNA is needed to move on to library prep.
The DNA can be stored at 4°C for up to four weeks, or -80°C indefinitely.
Part 6: Manual SQK-LSK114 Library Prep and Sequencing ( , including reloads) 6h 0m 0s, including reloads)
Note: Library prep can also be done on the Hamilton NGS Star and can process 48 samples in ~4 hours using the HAMILTON NGST STAR Oxford Nanopore SQK-LSK114 Library preparation SOP: https://dx.doi.org/10.17504/protocols.io.n2bvj36mnlk5/v1
Note: Reagents are from Oxford Nanopore Ligation Sequencing Kit V14 (SQK-LSK114). Library prep is following the standard Oxford Nanopore Ligation Sequencing DNA V14 (SQK-LSK114) protocol with minor tweaks: https://community.nanoporetech.com/docs/prepare/library_prep_protocols/genomic-dna-by-ligation-sqk-lsk114/v/gde_9161_v114_revo_29jun2022?devices=promethion
Minor tweaks:
2.5µgDNA library in48µL(Part A Step 2)- Add
45µLof AMPure XP beads (Part B Step 10) - Use Short Fragment Buffer (Part B Step 14)
- Incubate for
0h 20m 0sat37°Cand 300-450 x g (Part B Step 18)
A. DNA Repair and End-Prep
- Place all the necessary reagents
On iceto thaw and the Agencourt AMPure XP beads out atRoom temperature. - Prepare the following in a 0.2mL thin-walled PCR tube:
48µLDNA (input2.5µg, this might be over48µLbut that is fine. Adjust the amount of beads to match the total volume of this mixture (sample + buffers/enzymes))
Note: The DNA mass should be at least 1.15µg. However, this protocol is optimized for 2.5µg of starting input. Do not exceed 168µL of sample + 12µL of the buffers/enzymes added (max. total volume of 180µL).
3.5µLNEBNext FFPE DNA Repair Buffer (vortex and spin down)3.5µLUltra II End-Prep Reaction Buffer (vortex and spin down)3µLUltra II End-Prep Enzyme Mix (do not vortex, spin down)2µLNEBNext FFPE DNA Repair Mix (do not vortex, spin down)
Note: In our experience, AMPure XP beads and Short Fragment Buffer perform best when thawed and used at Room temperature rather than On ice.
-
Mix thoroughly by gently flicking the tube or very gently pipetting up and down 10x, and then spin down.
-
Using a Thermocycler, incubate samples at
20°Cfor0h 30m 0sand then65°C
for 0h 5m 0s and then 4°C for infinity.
Note: Start and pause Thermocycler to allow lid to come to 85°C before putting samples in.
-
Resuspend the AMPure XP beads by vortexing.
-
Transfer DNA samples to clean 1.5mL Eppendorf DNA LoBind tube.
-
Add
60µL(or equivalent volume to the previous sample-end-prep-mix, see step 2) of resuspended beads to the reaction and mix by flicking the tube 10x. Do not pipette mix here as beads may clump around the pipette tip. -
Incubate on a rocking laboratory shaker or hula mixer for
0h 5m 0satRoom temperature. -
Prepare
500µLper sample of fresh 80% ethanol in nuclease-free water. -
Spin down and pellet sample on magnet until eluate is clear and colorless, about
0h 2m 0s -
Keep the tube on the magnet and pipette off the supernatant.
Note: Can retain if needed just in case the final elution quantification is uncharacteristically low.
- With the samples remaining on the magnet, wash the beads with
200µLof 80% EtOH, pipetting on the opposite wall making sure not to disturb the pellet. Once EtOH has been added to all samples, immediately remove and discard the EtOH.
Note: The goal here is to make sure the beads are fully covered. If initial volume of beads was significantly higher than 60µL, more EtOH may be required.
-
Repeat step 12.
-
Spin down and place the tube back on the magnet, pipetting off any residual EtOH.
-
Allow to dry for ~
0h 0m 30sbut do not overdry to the point of cracking. -
Remove the tube from the magnetic rack and resuspend the pellet in
62µLnuclease-free water. Incubate on a rocking laboratory shaker or hula mixer for0h 3m 0satRoom temperature. If not quantifying the sample post-DNA repair and end-prep, user can resuspend the pellet in60µLwater. -
Spin down and pellet the samples on a magnet until eluate is clear and colorless.
-
Remove and retain
62µLof eluate into a clean 1.5mL Eppendorf DNA LoBind tube. -
Optional: Quantify
1µLof sample on the Qubit.
Note: It is possible to store samples at 4°C at this step if necessary.
B. Adapter Ligation and Clean-Up
- Spin down the Ligation Adapter (LA) and NEBNext Quick T4 DNA Ligase, then return to
On ice.
Note: Do not allow LA or Quick T4 to remain at Room temperature for too long. Since LA and Quick T4 do not freeze at -20°C, it is possible to leave them in the freezer until needed.
-
Thaw Ligation Buffer (LNB) at
Room temperature, mix by pipetting up and down (vortexing is ineffective due to viscosity), and placeOn ice. -
Thaw Elution Buffer (EB) and Short Fragment Buffer (SFB) at
Room temperature, mix by vortexing, spin down, and placeOn ice. -
In a 1.5mL Eppendorf DNA LoBind tube, mix the following in order:
60µLDNA sample25µLLNB10µLQuick T45µLLA
-
Mix by gently pipetting and spin down.
-
Incubate the reaction for
0h 30m 0satRoom temperature. -
During this time, put flow cells out at
Room temperature. -
Resuspend AMPure beads by vortexing.
-
Add
45µLof resuspended beads to the reaction and mix by flicking. -
Incubate on a rocking laboratory shaker or hula mixer for
0h 5m 0satRoom temperature. -
Spin down sample and pellet on magnet.
-
Keeping tube on magnet, pipette off the supernatant.
Note: Can retain supernatant if needed just in case the final elution quantification is uncharacteristically low.
-
Wash the beads with
250µLSFB, remove from magnet and flick to resuspend, spin down and re-pellet on magnet, and then remove and discard supernatant. -
Repeat step 14.
-
Spin down and place the tube back on magnet, pipetting off any residual supernatant.
-
Allow to dry for ~
0h 0m 30s, but do not overdry to the point of cracking. -
Remove the tube from magnet and resuspend pellet in
26µLEB, spin down, and incubate for0h 20m 0sat37°Cand 300-450 x g. -
During this time, QC the flow cells.
Note: Wait at least 0h 20m 0s after taking out the flow cells to allow them to reach Room temperature before loading onto the PromethION to avoid condensation formation.
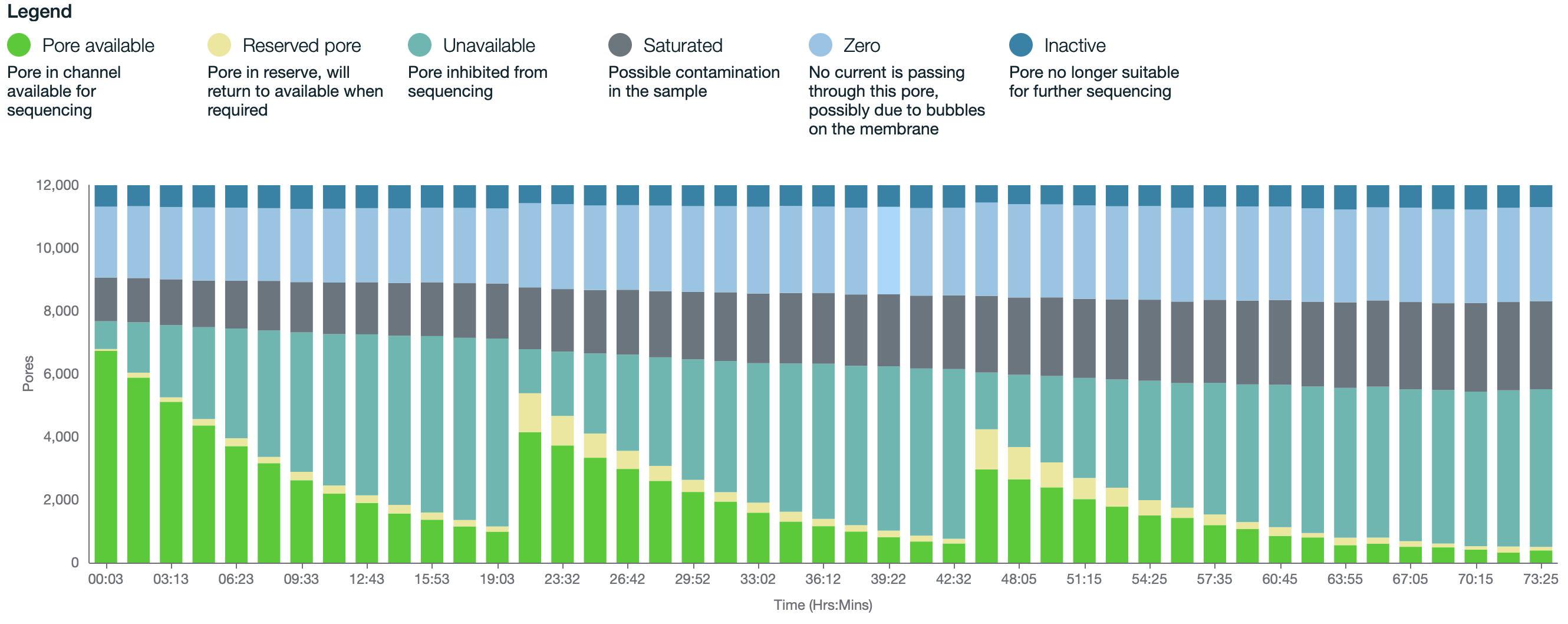
Note: If trying to reach 30x coverage, it is recommended to only use flow cells with >7000 pores.
-
Pellet the beads on a magnet until eluate is clear and colorless.
-
Remove and retain
26µLof eluate into a clean 1.5mL Eppendorf DNA LoBind tube, being careful not to aspirate any beads (this is the DNA library). -
Quantify
1µLof sample on the Qubit. -
Re-prep library from Part 6 if
<20fmol. Calculate the mass (ng) of DNA need to achieve this based on averaging the size of DNA from the Femto Pulse traces using https://nebiocalculator.neb.com/#!/dsdnaamt. This should allow a coverage of 30x over three loads. If your sample differs from this size, recalculate based on 10-20 fmol per load.
Note: For example, 740ng would be enough for three loads of 370ngeach, which is calculated for 20 fmol of DNA averaging at 30 kb. If necessary, user can load a higher fmol on the first load and lower fmol on the next two loads.
- Keep libraries
On iceuntil ready to load on flow cell.
C. Priming and Loading R10 Flow Cell
Note: This kit is only compatible with R10.4.1 flow cells (FLO-PRO114M).
-
Thaw Sequencing Buffer (SB), Library Solution (LIS) or Library Beads (LIB), Flow Cell Tether (FCT), and Flow Cell Flush (FCF) at
Room temperature, vortex, and spin down. -
Add
30µLof thawed and mixed FCT directly to tube of thawed and mixed FCF and vortex. Alternatively, in a new tube, add30µLof thawed and mixed FCT to1170µLof thawed and mixed FCF and vortex. This is your priming mix.
Note: You can make a Master-Priming mix as well: add 25µL of thawed and mixed FCT to 975µL of thawed and mixed FCT and vortex. Multiply by the number of samples and adjust for a little more volume than needed.
-
Rotate valve to reveal inlet port 1 on the flow cell, set P1000 pipette to
200µLand use the dial to draw back a small amount of volume to remove any air bubbles (usually about 20-30 μL, just until a small volume of buffer enters the pipette tip). -
Flush
500µLof priming mix into inlet port 1 of the flow cell, being extremely careful to avoid the introduction of air bubbles at the end. -
Wait
0h 5m 0s -
During this time, make up your DNA library to
32µLat20fmolusing EB. -
Prepare the library mix for loading:
100µLSB68µLLIS or LIB32µLDNA library (20fmol)
-
Repeat steps 3 and 4.
-
Gently pipette mix the prepared library mix right before loading.
-
Load
200µLof the library mix into inlet port 1 on the flow cell directly after the 2nd load of priming mix. -
Close valve to seal inlet port and close PromethION door.
-
Wait at least
0h 10m 0s(better up to1h 0m 0s) then initiate sequencing (see step 7.4 ). -
Ideally, the library quants yielded allow for 3
20fmolloads, the latter 2 loaded approximately after 24 and 48 hours. However this will vary slightly depending on pore usage, data generated, as well as other factors (i.e., if after 24 hours there are still 3000+ pores then the sample does not need to be reloaded until 48 hours).
Note: See step 7.5 Recovery of Library if you don't have 3 times 20fmol loads in total.
- To wash and reload a flow cell, begin by thawing Wash Mix (WMX)
On iceand Wash Diluent (DIL) atRoom temperature
Note: DIL should be vortexed. WMX should NOT be vortexed, only spun.
-
In a new tube, add
2µLWMX to398µLDIL and pipette mix. This is your flow cell wash mix. -
Pause the PromethION runs and export .pdf run reports.
-
Rotate the inlet port 1 cover to reveal inlet port 1.
-
Using a P1000, insert tip into inlet port 1 and draw back a small volume using the wheel to remove any air bubbles (usually around 20-30 μL, just until a small volume enters the pipette tip).
-
Load
400µLflow cell wash mix into inlet port 1, avoiding any introduction of air. -
Wait
1h 0m 0s -
With inlet port 1 closed, remove waste from port 2 or 3 (
920-950µLare expected) -
Repeat priming steps and reload samples (steps 1 - 13).
D. Starting a Run on PromethION
- Once library is loaded and the time has passed in step 12, navigate to 'Start sequencing'
Note: be sure that there is at least 2TB of data storage capacity per sample which can be found under 'Host settings'
- Select proper flow cell position and enter experiment name and sample ID name
Note : Type of flow cell should be automatically detected. Do not change this setting.
-
Select kit "Ligation Sequencing Kit SQK-LSK114"
-
Chose Run limit: "75 hours duration" (72 hours of running time plus additional time for paused wash and reloading steps) with a minimum read length of "200 bp" and Adaptive Sampling "off"
-
Chose basecalling options: Basecalling off is standard.
Note: We typically basecall after the run with Dorado (the advantage being with storage capacity and accuracy of the data). If selecting basecalling during the run, be sure to check if PromethION can handle the capacity (i.e. Fast basecalling can keep up with the speed of data acquisition on most Nanopore platforms. High Accuracy basecalling cannot.)
-
Chose data output files: ".POD5" for raw reads
-
After final review, start the sequencing process
Note: Check after 0h 10m 0s if the sequencing run looks good and there are not any air bubbles etc. If there is an issue with the flow cell, recover the library immediately with Step E.
E. Recovery of Library (optional)
- If there is not enough library for 3 complete 20 fmol loads, recovery of the previous library is possible. Before you doing the steps in 7.3.13 : rotate the inlet port 1 and remove
220µLvolume (last library load; a little bit yellow) and put that immediately on ice. - Add
20µLof SB and pipette mix. - Add any additional amount or rest of your library.
- Continue with steps 7.3.13 and mix instead of your library at step 7.3.10 with the total volume (
>200µL).
Note: You can not overload the flow cell if it will be more than 20fmol in the end.
F. Removing Flow Cells
-
After runs are complete, if basecalling was selected, make sure the basecalling shows 100% before removing flow cells.
-
Remove flow cells from PromethION and place aside to send back to Nanopore for recycling.