PacBio Iso-Seq Preparation for Sequel II Systems
Emil Gustavsson
PacBio Iso-Seq preparation
cDNA synthesis
cDNA amplification
LD PCR
amplified cDNA purification
cDNA purification
ASAPCRN
Abstract
The Sequel Systems generate long reads that are well-suited for characterizing fulllength transcripts produced from high-quality RNA samples. This document describes a method to construct Iso-Seq SMRTbell® libraries for sequencing on both systems allowing detection of full-length transcripts.
This protocol describes how to perform PacBio targeted Iso-Seq
Attachments
Steps
cDNA synthesis using SMARTer™ PCR cDNA Synthesis Kit First-Strand cDNA Synthesis
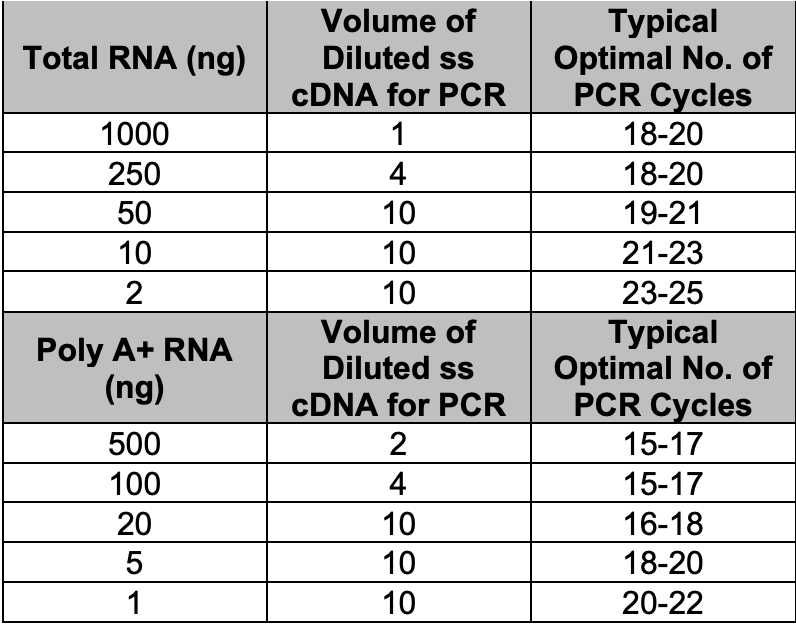
For each sample and Control Mouse Liver Total RNA, combine the following reagents in separate 0.5 ml reaction tubes:
| A | B |
|---|---|
| 1–3.5 μl | RNA (1 ng–1 μg of poly A+ RNA or 2 ng–1 μg total RNA) |
| 1 μl | 3’ SMART CDS Primer II A (12 μM) |
| x μl | Deionized H2O |
| 4.5 μl | Total Volume |
Mix contents and spin the tubes briefly in a microcentrifuge.
Incubate the tubes at 72°C in a hot-lid thermal cycler for 0h 3m 0s, then reduce the temperature to 42°C for 0h 2m 0s.
Prepare a Master Mix for all reaction tubes at room temperature by combining the following reagents in the order shown:
| A | B |
|---|---|
| 2 μl | 5X First-Strand Buffer |
| 0.25 μl | DTT (100 mM) |
| 1 μl | dNTP Mix (10 mM ) |
| 1 μl | SMARTer II A Oligonucleotide (12 μM) |
| 0.25 μl | RNase Inhibitor |
| 1 μl | SMARTScribe Reverse Transcriptase (100 U) |
| 5.5 μl | Total Volume added per reaction |
Aliquot 5.5µL into each reaction tube. Mix the contents of the tubes by gently pipetting and spin the tubes briefly to collect the contents at the bottom.
Incubate the tubes at 42°C for 1h 30m 0s.
Terminate the reaction by heating the tubes at 70°C for 0h 10m 0s.
Dilute the first-strand reaction product by adding the appropriate volume of TE buffer (10millimolar (mM), 0.1millimolar (mM)):
Add 40µL if you used total RNA as the starting material.
Add 190µL if you used more than 0.2µg as the starting material.
Add 90µL if you used less than 0.2µg as the starting material.
cDNA Amplification by LD PCR
For each reaction, aliquot the appropriate volume (see Table I, above) of each diluted first-strand cDNA into a labeled 0.5 ml reaction tube. If necessary, add deionized H2O to adjust the volume to 10µL.
Prepare a PCR Master Mix for all reactions, plus one additional reaction. Combine the following reagents in the order shown:
| A | B |
|---|---|
| 74 μl | Deionized H2O |
| 10 μl | 10X Advantage 2 PCR Buffer |
| 2 μl | 50X dNTP Mix (10 mM) |
| 2 μl | 5’ PCR Primer II A (12 μM) |
| 2 μl | 50X Advantage 2 Polymerase Mix |
| 90 μl | Total Volume per reaction |
Mix well by vortexing and spin the tube briefly in a microcentrifuge.
Aliquot 90µL into each tube from Step 11.
Cap the tube. Commence thermal cycling using the following program:
• 95°C 0h 1m 0s
• X number of cycles (consult Table 1).
• 95°C 0h 0m 15s
• 95°C 0h 0m 30s
• 95°C 0h 3m 0s
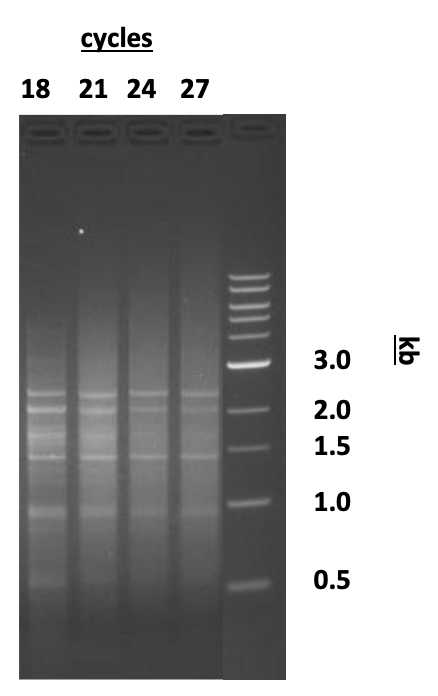
Subject each tube from step 9 to 15 cycles, then pause the program. Transfer 30µL to a second reaction tube labelled “Optimization”. Store the “Experimental” tubes at 4°C. Using the Tester PCR tube, determine the optimal number of PCR cycles:
Transfer 5µL from the 15 cycle PCR reaction tube to a clean microcentrifuge tube (for agarose/EtBr gel analysis).
Return the Optimization tubes to the thermal cycler. Run three additional cycles (for a total of 18) with the remaining 25µL.
Transfer 5µL from the 18 cycle PCR reaction tube to a clean microcentrifuge tube (for agarose/EtBr gel analysis).
Run three additional cycles (for a total of 21) with the remaining 20µL.
Transfer 5µL from the 21 cycle PCR to a clean microcentrifuge tube (for agarose/EtBr gel analysis).
Run three additional cycles (for a total of 24) with the remaining 15µL.
Transfer 5µL from the 24 cycle PCR to a clean microcentrifuge tube (for agarose/EtBr gel analysis).
Run three additional cycles (for a total of 27) with the remaining 10µL.
Electrophorese each 5µL aliquot of the PCR reaction alongside 0.1µg on a 1.2% agarose/EtBr gel in 1X TAE buffer. Determine the optimal number of cycles required for each experimental and control sample.
Retrieve the 15 cycle Experimental PCR tubes from 4°C, return them to the thermal cycler, and subject them to additional cycles, if necessary, until you reach the optimal number.
Add 2µL to each tube to terminate the reaction.
Purification of Amplified cDNA
Add 1X AMPure PB beads to the amplified cDNA.
Mix by tapping the LoBind tube until the sample is homogeneous.
Incubate at 4Room temperature for 0h 10m 0s.
Place on magnetic rack until solution clears. Remove and discard supernatant.
With the tube still on magnet, add 200µL to the tube containing beads plus DNA (1/2).
Remove and discard 70% ethanol (1/2).
With the tube still on magnet, add 200µL to the tube containing beads plus DNA (2/2).
Remove and discard 70% ethanol (2/2).
Let beads air dry for 0h 1m 0s.
Add 27µL and remove the tube from the magnet. Mix by tapping the tube until the sample is homogeneous. Then incubate at 4Room temperature for 0h 2m 0s.
Place back on magnet. When the solution clears, remove 25µL into new 1.5 mL LoBind tube.
Determine concentration using Qubit device or similar quantification assay.
Run 1µL on Agilent DNA 12000 chip according to manufacturer’s instructions.
The captured cDNA is now ready for SMRTbell library construction.
Repair DNA Damage
In a LoBind microcentrifuge tube, add the following reagents:
| A | B |
|---|---|
| X μl | cDNA for 500 ng |
| 7 μl | DNA Prep Buffer |
| 0.6 μl | NAD |
| 2 μl | DNA Damage Repair Mix v2 |
| Up to 57 μl | H2O |
| 57 μl | Total Volume per reaction |
Pipette mix 10 times. It is important to mix well. Perform a quick spin to collect all liquid from the sides of the tube.
Place in a thermocycler and run the following program:
• 37°C 0h 30m 0s
• Hold at 4°C
End Repair/A-Tailing
In a LoBind microcentrifuge tube, add the following reagents:
| A | B |
|---|---|
| 57 μl | Reaction Mix from previous step |
| 3 μl | End Prep Mix |
| 60 μl | Total Volume per reaction |
Pipette mix 10 times. It is important to mix well. Perform a quick spin to collect all liquid from the sides of the tube.
Place in a thermocycler and run the following program:
• 20°C 0h 30m 0s
• 65°C 0h 20m 0s
• Hold at 4°C
Overhang Adapter Ligation
Add the following directly to reaction mix from previous step:
| A | B |
|---|---|
| 60 μl | Reaction Mix from Previous Step |
| 3 μl | Overhang Adapter v3 |
| 30 μl | Ligation Mix |
| 1 μl | Ligation Enhancer |
| 1 μl | Ligation Additive |
| 95 μl | Total Volume per reaction |
Pipette mix 10 times. It is important to mix well. Perform a quick spin to collect all liquid from the sides of the tube.
Place in a thermocycler and run the following program:
• 20°C 1h 0m 0s
• Hold at 4°C
Purification of cDNA
Add 1X AMPure PB beads to the amplified cDNA.
Mix by tapping the LoBind tube until the sample is homogeneous.
Incubate at 4Room temperature for 0h 10m 0s.
Place on magnetic rack until solution clears. Remove and discard supernatant.
With the tube still on magnet, add 200µL to the tube containing beads plus DNA (1/2).
Remove and discard 70% ethanol (1/2).
With the tube still on magnet, add 200µL to the tube containing beads plus DNA (2/2).
Remove and discard 70% ethanol (2/2).
Remove ethanol.
Check for any remaining droplets in the tube. If droplets are present spin down down and place tube back on magnetic rack and pipette of any remaining ethanol.
Let tube air dry for 0h 1m 0s.
Add 30µL and remove the tube from the magnet. Mix by tapping the tube until the sample is homogeneous. Then incubate at 4Room temperature for 0h 2m 0s.
Place back on magnet. When the solution clears, remove 30µL into new 1.5 mL LoBind tube.
Purification of cDNA library
Perform two rounds of Ampure PB bead clean up in the "Purification of cDNA" section.
Prepare for Sequencing
Follow the SMRT Link Sample Setup v8.0 (or higher) instructions for preparing the sample for sequencing on the Sequel II System.