PROTOCOL FOR PIGMENT CONTENT QUANTIFICATION IN HERBACEOUS COVERS: SAMPLING AND ANALYSIS
Rosario R. Gonzalez-Cascon, Maria Pilar Martin
Abstract
The scope of the protocol is the quantification of pigments in pasture samples in the context of proximal and remote sensing-based studies. The protocol describes a standard procedure for the destructive sampling of mixed pasture covers for the estimation of biophysical variables (biomass, leaf area index, canopy water content, specific leaf area, specific leaf weight), pigments, and nutrients without discrimination by species or functional types and the laboratory procedure for the samples pretreatment and quantification of pigments contents (Chlorophyll a, Chlorophyll b, and carotenoids) in the collected samples. The procedure allows the quantification of pigments in herbaceous covers per fresh or dry mass as well as per sampling area. The determination of pigments is performed by a cold extraction procedure with 80% v/v aqueous acetone followed by a spectrophotometric determination.
Before start
Steps
1. FIELD SAMPLING PROTOCOL
The filed protocol is designed to sample 2525 m permanent plots randomly located inside the footprint of an Eddy covariance tower installed in a tree-grass ecosystem. Inside each plot at least two 11 m quadrant are used for destructive herbaceous layer sampling. The number of 1*1 m quadrants located in and outside of the tree crown influence area reflects the tree cover percentage of the tree-grass ecosystem.
EQUIPMENT AND MATERIALS
-
Supplies
-
Large sampling quadrats (1x1m)
-
Small sampling quadrats (25x25cm)
-
Adhesive tape for labeling
-
Scissors and cutters
-
Plastic zip sampling bags
-
Permanent markers
-
Disposable gloves
-
Portable cooler + ice bricks
-
Expanded Polypropylene (EPP) thermal box filled dry ice pellets
-
Aluminum foil
-
Equipment
-
Tripod
-
Bubble level
-
Compass
-
Selfie stick or rigid stick for the tripod
-
Cell phone with Epicollet5 app installed (https://five.epicollect.net/)
-
Electronic field scale
SAMPLING STEPS
1x1m sampling quadrants localization within the 25x25 m reference plots. At least two quadrants will be located in homogeneous patches visually identify within the reference plot in areas that represent the variability of the grass cover.
- 1x1m sampling quadrants localization within the 25x25 m reference plots. At least two quadrants will be located in homogeneous patches visually identify within the reference plot in areas that represent the variability of the herbaceous cover of the reference plot.
- Metadata collection of 11 m quadrants using the Epicollet5 app. With a pre-designed project within the Epicollet5 app acquire the field characterization of each 11 m quadrant: GPS location, sampling date and time, general picture of the quadrant, plot number, quadrant type, operator name, and field observations. The picture is taken with the cell phone from a fixed position (North quadrant side) at a fix height with help of the tripod and self-stick.
- 25x25 cm biophysical and chlorophyll sampling quadrats location inside 1x1m (Figure 1).
- Destructive grass collection biophysical variables ( BIO-sample ).
- Destructive grass collection for pigment determination ( Chl-sample ).
- Weighting and preserving the samples.
Destructive grass collection for BIO-samples
Two samples are taken in the 25*25 cm quadrant: pasture and litterfall.
Label the sampling plastic zip bags with the following information:
- Plot code
- Quadrant number
- Sample type:
- Open grass (OG)
- Below canopy grass (BCG) (when the sample is located below or under the influence of a tree crown)
- Open litterfall (OL)
- Below canopy Litterfall (BCL)
- Date
- Time
- Soil: wet OR dry
- Plant superficial water: YES or NOT
- Collector’s name or ID
-
Locate the 2525 quadrant inside the 11 m quadrant for biophysical sampling (Figure 1).
-
Collect the pasture sample. Pull/cut live grass (green or dried), flowers, and spikes until no vegetation remains inside the quadrant introduce them in the plastic zip bag. Do not include roots, soil, insects, etc.
-
Collect the litterfall: introduce in the plastic zip bag all dead vegetal debris (waffle, dead grass, little branches, seeds, etc.). Please do not include, soil, insects, etc. neither dry (dead looking) rooted plants.
-
Carefully close the plastic zip bags and avoid punctures and breaks in the bag otherwise you could lose part of sample or changes in its moisture.
-
Introduce the samples in a big plastic bag inside the portable cooler with ice bricks (the sample bags should never be in contact with the ice bricks). Bring the cooler to the base camp to weigh the samples.
Destructive grass collection for Chl-samples
One pasture sample is collected in the 25*25 cm quadrant.
- Use disposables gloves for pigment pasture collection.
- Label the zip plastic bag with:
- Plot code
- Quadrant number
- Sample type
- Open grass (OG)
- Below canopy grass (BCG) (when the sample is located below or under the influence of a tree crown)
- Date
- Time
- Collector’s name
- Collect only fully green plants or the green parts in case a plant have green and not green parts. Do not include in the sample bag non-photosynthetic parts of the plant such as roots, flowers, spikes, etc.
- Carefully close the plastic zip bag. Introduce them inside the portable cooler. Bring the cooler to the base camp as quickly as possible to weight and freeze the sample.
Weighing and preserving samples procedure
- BIO-sample:
- Place and level the electronic scale in the base camp.
- Weigh each sample bag.
- Write the weight in the plastic zip bag using a permanent marker.
- Return the sample to the field cooler container.
- Chl-sample:
- Weigh each sample bag.
- Write the weight in the plastic zip bag using a permanent marker.
- TAKE A PICTURE of the sample ZIP PLASTIC BAG
- Introduce a paper label sample identification inside the bag
- Wrap the plastic zip bags with aluminum foil. Label aluminum foil.
- Froze immediately the sample in dry ice or liquid nitrogen
Samples for pigment determination are transported in a EPP thermal box filled with dry ice pellets until the laboratory. The samples in the laboratory are stored immediately in the freezer.
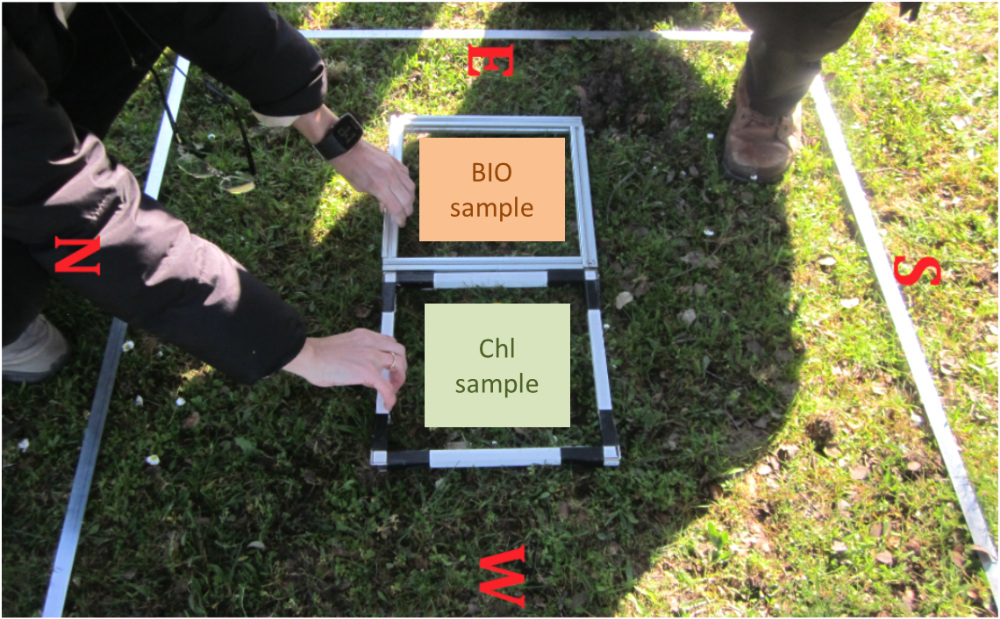
Figure 1: Field set-up for destructive pasture sampling inside the 1*1 m quadrant.
2. LABORATORY SAMPLES PRE-TREATMENT
Objective
- Avoiding pigment degradation during storage.
- Assure a homogenization of the sample. Samples pre-grinding is necessary to homogenize a sample composed from a mix of several grass species. For this purpose, frozen pasture samples are pre-ground in a mill with dry ice to avoid pigment degradation.
EQUIPMENT AND MATERIALS
1. Supplies
- Dry ice pellets
- Disposable gloves
- L7651 Avery labels
- Aluminum foil
- Spatula, stainless steel spoon
- Scissors
- Polyethylene plastic containers (50 or 100 ml)
- Ethanol
- Cotton swabs
- Permanent marker
- Black EPP thermal box
2. Equipment
-
Mill
-
Freezer
-
Compressed air PROCEDURE
-
Pasture samples are preserved in the freezer at -20ºC.
-
Label the plastic sample containers before grinding. Depending on sample size, 50 ml or 100 ml plastic containers are used. Label the lid with the laboratory registration number and the bottle with the following sample identification parameters:
- Laboratory register number
- Plot number
- Pasture type
- Sampling quadrant number
- Sampling date
- Transport the frozen samples from the freezer to the dark room in a black box filled with dry ice.
- Put the scissors, spatula and sample containers into the dry ice box. Cool the mill by putting some dry ice pellets into the mill hollow and cover it.
- Take the pasture sample out of the plastic bag over a small tray covered with aluminum foil with ice pellets. Pre-cut the pasture leaves quickly with the cold scissors. Introduce the sample into the mill cavity.
- Add dry ice pellets and grind the mixture until achieving a homogeneous aspect (Figure 2). IMPORTANT : the entire pasture material collected in the field from the 25*25 cm quadrant should be ground to assure a complete homogenization of the sample.
- Put a pellet of dry ice in the bottom of the sample container ant transfer the ground sample with the cold spatula into the labelled container.
- Cover the sample container with aluminum foil and return the sample to storage.
- Dry (air dry) the plastic field bags used to store the sample in the field, clean with compressed air, weight (laboratory scale ±0.00 precision) and annotate the bag mass (g). This step is necessary to calculate the pasture fresh and dry mass sampled inside the 25*25 cm quadrant.

Figure 2: Mill (left), pre-cooled mill for grinding (center) and aspect of a homogenized pasture sample after grinding (right).
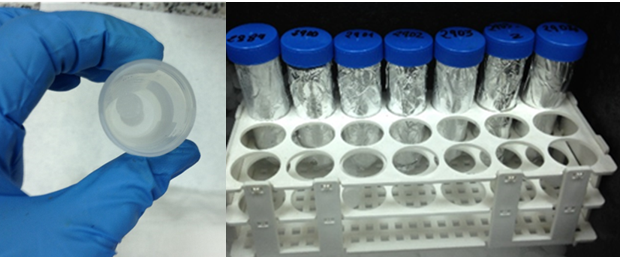
Figure 3: Dry ice pellet in the bottom off a sample container (left), homogenized pasture samples after grinding (right).

Figure 4: Pasture sample from 25*25 cm2 quadrant (left), homogenized sample in dry ice (right).
3. PIGMENT EXTRACTION PROCEDURE WITH 80% AQUEOUS ACETONE
Objective
Quantification of Chlorophyll a, Chlorophyll b, and carotenoids contents from the homogenized grass sample by a cold extraction procedure with 80% v/v aqueous acetone followed by a spectrophotometric pigment determination using Wellburn equations (1994).
EQUIPMENT, MATERIALS AND REAGENTS
Supplies
-
10 ml Glass reagent tubes (2/extract): one PIREX tube with screw cap for maceration and one glass tube for the spectrophotometric determination
-
2 ml Eppendorf (1/extract)
-
Stainless steel balls for the ball mill (≈3mm diameter, Retsch 22.455.0002)
-
Racks for the Eppendorf- and reagent tubes
-
Scissors, tweezers, microspatula
-
Paired quartz cuvettes (HELLMA®)
-
5 ml glass containers (2/sample) for dry mass determination
-
3 l plastic jar
-
Disposable gloves
-
Aluminum foil
-
Permanent marker
-
Black EPP box filled with crushed ice Reagents
-
Acetone Chromasolv® HPLC 99.8% (Sigma-Aldrich)
-
Deionized wate Equipment
-
Precision scale
-
Teflon top-dispenser (5 ml) with 2l glass bottle (Fortuna® Optifix® 5ml)
-
Desiccator
-
Stove (65ºC)
-
Ball mill with 2 adapter for 2ml eppendorf tubes (Techtnica Millmix 20)
-
Centrifuge
-
Eppendorf ThermoMixer® C
-
Spectrophotometer
PIGMENT EXTRACTION
Objective
Complete extraction of pigments from homogenized grass samples.
Laboratory analysis are conducted in a dark room. Use personal protective equipment (disposal gloves, safety glasses, and lab coat).
Laboratory preparations the day before extractions
-
Label the PIREX-, glass- and Eppendorf tubes.
-
Label the two 5 ml glass flasks for dry mass determination.
-
Print laboratory forms.
-
Put acetone and water into the refrigerator.
-
Put stainless steel 3 mm balls into the freezer. Pigment extraction procedure
-
Switch on the scale and stove (65ºC).
-
Fill the EPP box with crushed ice. Put inside the rack with the 2ml eppendorf tubes and the two boxes for the ball mill.
-
Prepare 500 ml solvent 80% acetone (v/v) with two graduated cylinders: 400 ml acetone + 100 ml water.
-
Transfer the solvent to the glass bottle with Teflon top-dispenser, drain it, and maintain the bottle cold by putting it into a plastic jar filled with crushed ice.
-
Prepare a reagent glass filled with crushed ice in which to keep the sample container cold during the weighting process.
-
Take the first sample out of the freezer, put it into the reagent glass, homogenize the sample with a spatula, and weight 40-60 mg pre-ground pasture sample into 2 ml Eppendorf tubes. Introduce two 3 mm balls in each Eppendorf tube. Add 1ml 80% acetone and put it into the ball mill box (EPP black box). Make four extractions per sample.
-
Prepare two blanks by adding 1 ml acetone into an Eppendorf tube.
-
Weight two replicates of approximate 1 g in a 5 ml glass flask for dry weight determination, introduce them into the stove, and return the sample to storage (65ºC 24 hours).
-
Repeat the process with the next sample.
-
Switch on the ball mill, place the two ball mill boxes, and fix it. Grind the samples for three minutes at 25 rpm.
-
Transfer quantitatively the pasture extract to a 10 ml PIREX reagent glass by washing the Eppendorf tube 3-4 times with 1 ml aqueous acetone and completing to a final volume of 8 ml. Take the two balls out of the PIREX reagent glass by using a magnetic bar. IMPORTANT : work under fume hood and use a protective safety laboratory mask during the transfer process to prevent inhalation of acetone.
-
Let the final 8 ml pigment extract macerate for two hours in the darkness (inside the cold EPP black box, covered with aluminum foil). Agitate the extract 4-5 times during the maceration.
-
An alternative way for the cold maceration process is the use of an Eppendorf ThermoMixer® C. Cool the mixer to 5ºC for 40 minutes before extraction. Introduce the PIREX tubes with screw cups into the mixer. Agitate the 8 ml pigment extracts at 5ºC and 850 rpm during one hour (Figure 6).
-
Put the pigment extracts into centrifuge (take out the tube screws). Centrifuge the extracts three minutes at 3000 rpm. Decant the supernatant into a 10 ml glass reagent tube. Preserve the extracts into the EPP black box.

Figure 5: Cold 80% acetone bottle (left), boxes of the ball mill with 2 ml Eppendorf tubes (second left), ball mill (center), 8 ml pigment extracts prepared for maceration (right).
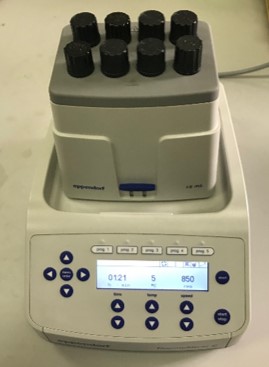
Figure 6: Cold maceration of pigment extracts in an Eppendorf ThermoMixer®.
SPECTROPHOTMETRIC MEASUREMENTS
Wellburn (1994) equations for pigment determination were developed specifically for spectrometric measurements in glass cuvettes with spectrophotometers of 0.1-05 nm or 1-4 nm resolution.
Absorption readings at three wavelengths are required for the calculations of Chlorophyll a (Chla), Chlorophyll b (Chlb), and carotenoids(Cx+c) with 80% v/v aqueous acetone: 663.2 nm, 646.8 nm and 470 nm. Additionally readings at 750 nm are performed for extract turbidity control.
Absorption readings should be inside the linear range of 0.300-0.750. In case of readings below this range, extracts should be repeated increasing the fresh extract weight. Over the range, extracts should be diluted with 80% acetone.
- Warm up the spectrophotometer 20 minutes before measurements. Beckman DU 530 UV/Vis allows the simultaneous measurements of four wavelengths.
- Material needed for the spectrophotometer measurements:
- Laboratory forms
- 80% acetone
- Laboratory tissues, glass container
- 5 ml paired glass cuvettes
- EPP cold box with pasture extracts
- Set the instrument to zero with 80% acetone. Measure the two blanks and extracts at the four wavelengths. Reset to zero with 80% acetone each 12 measurements.
- After analysis, clean all the glass material and put it in the stove for glassware. Dispose the plastic material and reagent/extract residues for chemical waste treatment.
CALCULATIONS
1. Pigment concentrations are calculated with the Wellbrun (1994) equations. In the case of spectrophotometer with 0.1-0.5 nm resolution and 80% v/v acetone solvent the equations are:
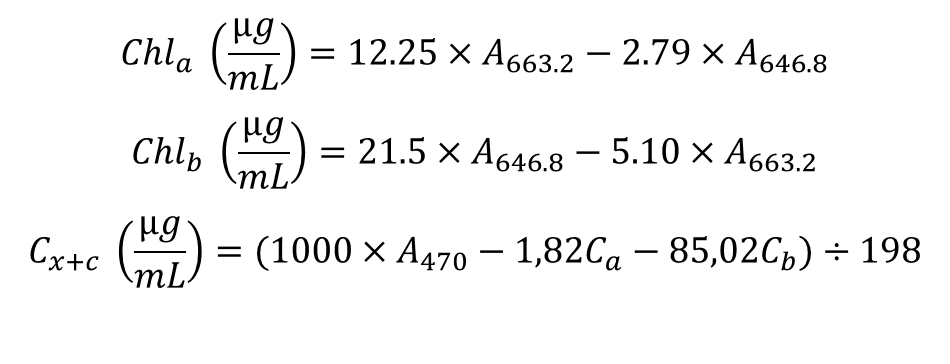
-
Pigment content per fresh mass is calculated as follows :
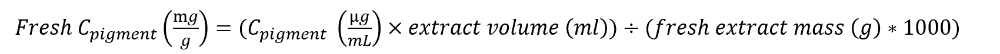
3. Pigment content in the pasture at 65ºC is calculated as follows:
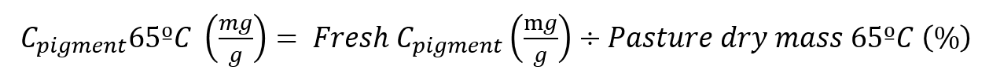
4. Pigment content per sampling surface unit (g/m2) is calculated as follow:
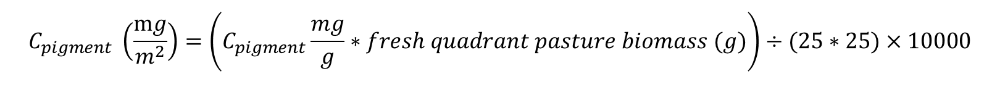
All the formulae are prepared in an Excel file. Inputs for the Excel file are the fresh pasture extraction mass (g), the four extract absorption readings, the pasture dry matter data (%), and the fresh quadrant pasture biomass (g).
QUALITY CHECKS
-
Extract turbidity: absorption readings at 750 nm greater than 0.005 indicate the presence of turbidity or the presence of colored interfering compounds at this wavelength.
-
Check the Chla/Chlb Too narrow Chla/Chlb ratios may indicate the presence of compounds that may interfere with the spectrophotometric measurements.
-
Check the variation coefficient of pigment concentrations in the four pasture sample replicates. Repeat extractions in case of failures during the extraction procedure.
Finally, dry an aliquot of the remaining pre-ground pasture sample at 65ºC for 48 hours for further carbon/nitrogen analysis (dry combustion analyser) or total nutrient analysis.
REFERENCES
Wellburn, A. R. (1994). 'The spectral determination of chlorophyll a and chlorophyll b, as well as total carotenoids, using various solvents with spectrophotometers of different resolution.' Journal of Plant Physiology 144(3): 307-313.

