Optogenetic Control of Subcellular Protein Location and Signaling in Vertebrate Embryos
Clare E. Buckley
Abstract
This chapter describes the use of optogenetic heterodimerization in single cells within whole-vertebrate embryos. This method allows the use of light to reversibly bind together an “anchor” protein and a “bait” protein. Proteins can therefore be directed to specific subcellular compartments, altering biological processes such as cell polarity and signaling. I detail methods for achieving transient expression of fusion proteins encoding the phytochrome heterodimerization system in early zebrafish embryos (Buckley et al., Dev Cell 36(1):117–126, 2016) and describe the imaging parameters used to achieve subcellular light patterning.
Steps
3.1 Plasmid Cloning (See Note 2)
Choose sequences encoding appropriate proteins to link to the PHYB “anchor” and PIF “bait.” For example, if you want to activate a GTPase then choose a membrane moiety such as CAAX to link to PHYB ( see Fig. 1a) and the relevant guanine exchange factor (GEF) to link to PIF ( see Note 9) .
![Fig. 1Phytochrome heterodimerization. (a) In this example, the phytochrome B (PHYB) “anchor” protein is linked to the membrane moiety CAAX and the phytochrome interaction factor (PIF) “bait” protein is linked to the EGFP fluorophore and the apical polarity protein Pard3. In the presence of phycocyanobilin chromophore (PCB), PHYB heterodimerizes with PIF under 633 nm illumination, therefore recruiting the fusion protein Pard3-EGFP-PIF6 from the cytoplasm to the membrane. After dimerization, PHYB and PIF proteins remain stably joined unless exposed to 740 nm light, which reverses the dimerization and sends Pard3-EGFP-PIF6 back into the cytoplasm. Both processes occur within a few seconds of light exposure. (b) “Binding” 633 nm light is patterned onto a subcellular region of the membrane and is overlaid by global “unbinding” 740 nm light. This results in the precise recruitment of Pard3-EGFP-PIF6 to the region of interest (adapted from Ref. [17] under the terms of the Creative Commons Attribution License (CC BY): http://creativecommons.org/licenses/by/4.0/) Fig. 1Phytochrome heterodimerization. (a) In this example, the phytochrome B (PHYB) “anchor” protein is linked to the membrane moiety CAAX and the phytochrome interaction factor (PIF) “bait” protein is linked to the EGFP fluorophore and the apical polarity protein Pard3. In the presence of phycocyanobilin chromophore (PCB), PHYB heterodimerizes with PIF under 633 nm illumination, therefore recruiting the fusion protein Pard3-EGFP-PIF6 from the cytoplasm to the membrane. After dimerization, PHYB and PIF proteins remain stably joined unless exposed to 740 nm light, which reverses the dimerization and sends Pard3-EGFP-PIF6 back into the cytoplasm. Both processes occur within a few seconds of light exposure. (b) “Binding” 633 nm light is patterned onto a subcellular region of the membrane and is overlaid by global “unbinding” 740 nm light. This results in the precise recruitment of Pard3-EGFP-PIF6 to the region of interest (adapted from Ref. [17] under the terms of the Creative Commons Attribution License (CC BY): http://creativecommons.org/licenses/by/4.0/)](https://static.yanyin.tech/literature_test/protocol_io_true/protocols.io.bp3jmqkn/370791_2_En_10_Fig1_HTML.png)
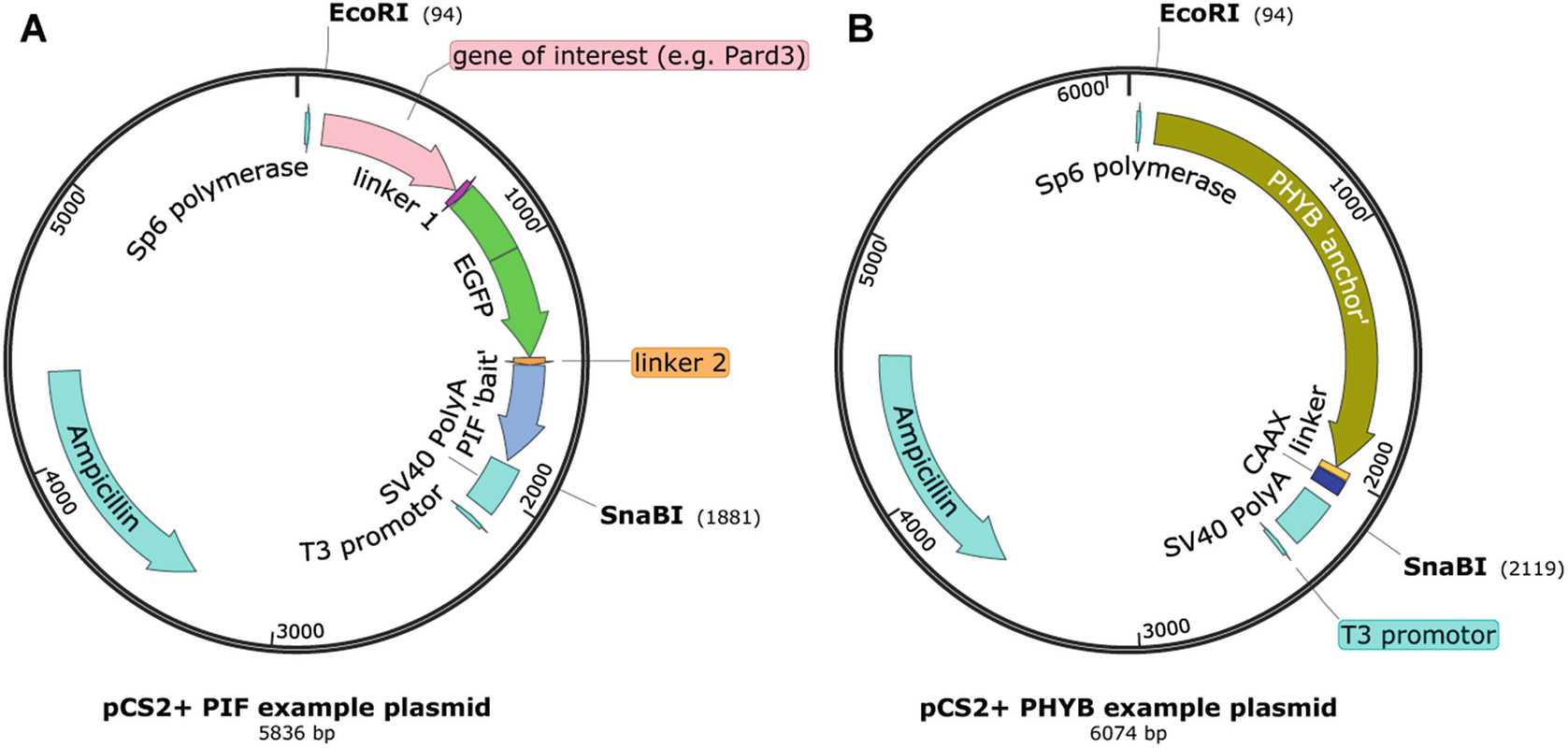
Design your PHYB- and PIF-containing plasmids using gene visualizing software. Your PHYB-linked protein should either be unlabeled or be labeled with a longer wavelength fluorophore ( see Note 10 ). The PHYB protein should only be tagged at the 3′ end, never at the 5′ end, which tends to inhibit function of the protein. You should use the shortened version of the PHYB protein (1–621 amino acids of Arabidopsis Gene ID 816394, codon optimized). Your PIF-linked protein should be labeled with a shorter wavelength fluorophore (green or blue-light responsive) to minimize background activation of the phytochrome system. PIF proteins can be tagged at either the 5′ or the 3′ end. Codon-optimized PIF6 (1–100 amino acids of Arabidopsis Gene ID 825382) or PIF3 (1–132 amino acids of Arabidopsis Gene ID 837479) can be used. Linkers (such as 10 amino acid polyglycine-serine) should be used between adjacent genes to allow for protein folding. Sequences for these fusion proteins should be incorporated into the pCS2+ vector plasmid between the restriction digest sites ECORI and SnaBI (Fig. 2; see Note 11 ).
If you are using Gibson assembly ( see Note 2 ), find templates encoding all the proteins that you want to combine into your plasmid and design and order primers to produce gene fragments with 30 base pair overlapping ends.
To prepare the vector ( steps 4 and 5 ), linearize the pCS2+ vector at two distinct sites using restriction enzyme digest: Incubate 1µg–5µg with ECORI enzyme and buffer to 20µL at 37°C for 2h 0m 0s–3h 0m 0s in a 1.5 μL microcentrifuge tube, as per the manufacturer’s instructions. Purify the linearized DNA in the tube using a DNA purification kit, as per the manufacturer’s instructions. Further digest this purified DNA as above, using SnaBI enzyme and buffer.
Run the whole digest onto a 1% electrophoresis gel: For a 1% gel, add 1 g of agarose to every 100 mL TAE buffer and melt in the microwave. In the fume hood, add 3µL and pour into the gel tank containing a comb. Leave until agarose is set. Add ladder to one well and DNA sample with the appropriate amount of gel-loading dye and water into 1–2 other wells. Run the electrophoresis machine as per the manufacturer’s instructions at around 100 V for 0h 20m 0s. Wearing a UV protective face shield and lab coat, visualize the gel on the UV transilluminator and cut out the appropriate weight of band using a razor blade. Place the gel containing the band into a 1.5 mL microcentrifuge tube and extract the DNA using a gel extraction kit, as per the manufacturer’s instructions.
Use the high-fidelity PCR kit to amplify your gene fragments using your primers and template DNA, as per the manufacturer’s instructions ( see Note 12 ).
Remove residual template from your PCR product either by Dpn1 digestion in the tube or via gel extraction, as above ( see Note 13 ).
Mix together your DNA fragments in 5µL with a molar ratio of 2:1 for insert fragment:vector backbone ( see Note 14 ). Add this to 15µL and incubate at 50°C for 1h 0m 0s.
Transform the Gibson assembly reaction into chemically competent cells: Thaw 50µL On ice. Still on ice, add 5µL to the cells and incubate for 0h 20m 0s–0h 30m 0s. Heat shock the cells at 42°C without shaking and then immediately transfer back to ice. Spread the cells onto two pre-warmed ampicillin-selective plates and incubate at 37°C.
Place the plate in the fridge in the morning. In the afternoon, pick 4–6 colonies and add each one to 4mL within a 14 mL Falcon tube. Shake at 37°C .
Mix 750µL with 750µL (50% glycerol, 25% nuclease-free water, 25% PBS) and store at -80°C.
Extract the plasmid DNA from 1.5 mLs of each E. coli suspension using a miniprep kit, as per the manufacturer’s instructions.
Carry out diagnostic restriction enzyme digests on a sample of each plasmid and run on an electrophoresis gel, as above, to check that you have made the expected plasmid. Send a sample for sequencing to confirm that there are no mutations.
Take the glycerol stock from the miniprep with the correct sequence and scrape some frozen cells with a sterile pipette tip. Drop this tip into 2mL in a 14 mL Falcon tube. Grow at 37°C in the shaker over the day. Add this 2 mL culture to 150mL in a sterile flask and grow at 37°C in the shaker . Extract the plasmid DNA using a midiprep kit, as per the manufacturer’s instructions.
3.2 mRNA Synthesis
Linearize the DNA template at a single site using Not1 restriction enzyme digest as in Section 3.1 'Plasmid Cloning', step 4 ( see Note 15 ).
Check that the linearization has occurred properly: Remove 1µL and run on an electrophoresis gel (as in Section 3.1 'Plasmid Cloning', step 5 ). Wearing a UV protective face shield and lab coat, visualize the gel on the UV transilluminator to determine whether you have a single band of the expected weight.
Ethanol precipitate the DNA ( see Note 16 ): Add 81µL to make up to 100 μL volume. Add 10µL and 400µL. Place at -20°C for at least 1h 0m 0s. Using the microcentrifuge, spin at 14000x g. Carefully remove and discard the supernatant, taking care not to remove the pelleted DNA at the bottom of the tube. Add 500µL and spin for 14000x g ( see Note 17 ). Carefully remove and discard the supernatant. Leave the top of the microcentrifuge open and dry the pellet at Room temperature. Resuspend the DNA pellet in 6µL.
Synthesize capped mRNA using the RNA transcription kit as per the manufacturer’s instructions ( see Note 18 ).
Purify the RNA using an RNA cleanup spin column kit as per the manufacturer’s instructions ( see Note 19 ).
Store in 0.5µL – 1µL aliquots at -80°C ( see Note 20 ).
3.3 mRNA Injection
Place a glass capillary inside the micropipette puller and adjust settings to pull an injection needle with an approximately 0.5 μm tip. The specific settings will vary by individual machine ( see Note 21 ) but an example for a Sutter P-87 machine is as follows: heat = 800, pull = 65, velocity = 55, and time = 210. Place the injection needles in a holding plate until ready to load ( see Note 22 ).
Mix the PhyB- and PIF-tagged mRNA and PCB ready for injection ( see Note 23 ). The exact concentration of RNA will depend on what you are injecting but a rough guide is to inject approximately five times more PHYB-encoding RNA than PIF-encoding RNA ( see Note 24 ). You should start the working concentration for PCB at 3millimolar (mM) and adjust it according to how much volume you are injecting and the outcome of your pilot experiments. The injection mix should ideally be used at 80Room temperature right away.
Under a dissecting microscope, use forceps to line up around 20 zebrafish embryos in the injection plate and remove excess water.
Under low-light conditions load the RNA/PCB mix into the injection needle using a fine loading tip.
Insert the needle into the micromanipulator attached to a microinjector and pump.
Use fine-tipped forceps to break the last 1–2 mm of the needle taper so that the tip is approximately 6 μm wide.
Working quickly to minimize exposure to light ( see Note 25 ), use a graticule to measure the drop size generated by the microinjector and adjust it to your required drop size ( see Note 26 26 ).
Use the micromanipulator to insert the needle into a single cell of an 8–32-cell-stage embryo and inject a single drop into the cell (Fig. 3).
Place the injected embryos in embryo medium in a petri dish and wrap the dish in aluminum foil to protect it from light. Incubate at 28.5°C until ready for imaging.
3.4 Embryo Mounting (See Note 27)
When the embryos are at the right stage for imaging use fine-tipped forceps to remove their chorions.
Using a glass pipette and bulb suck up the embryo in a small amount of water.
Drop the embryo into the melted low-melt-point agarose at 50°C–55°C ( see Note 28 ). Remove the remainder of the water in the pipette and suck up the embryo in agarose.
Place the embryo in a drop of agarose inside the imaging dish and orient with forceps under a dissecting microscope. Repeat with any other embryos for imaging.
Allow the agarose to set for around 0h 1m 0s and cover the embryos with embryo medium. If the embryos will be 16 h postfertilization (h.p.f.) or above during imaging then add tricaine to the embryo medium to prevent embryos from moving.
Wrap the imaging dish in aluminum foil to protect from light until imaging.
3.5 Live Imaging and Optogenetic Subcellular Localization of Proteins (See Note 29 and Figs. 4 and 5)
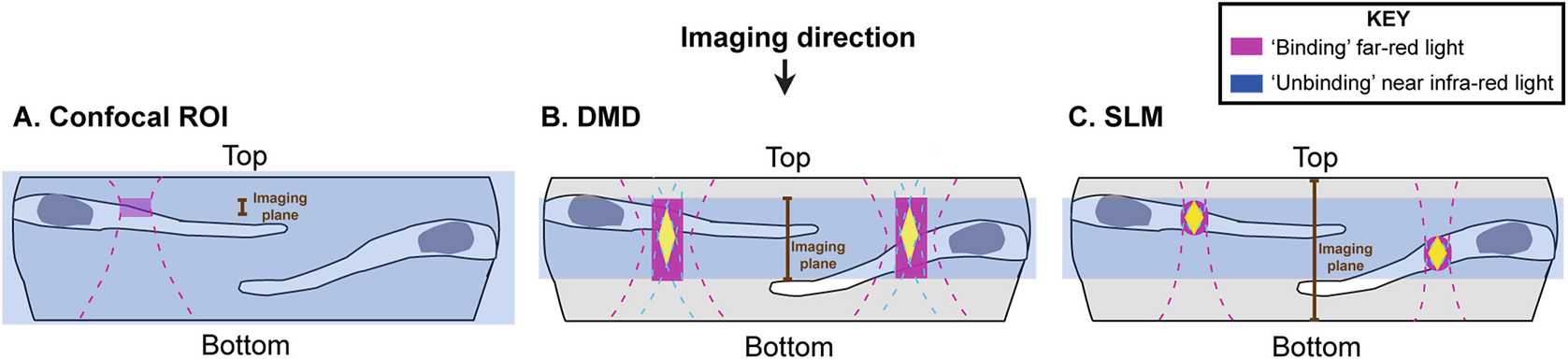
Ensure that the dark imaging chamber on the microscope is preheated to 28.5°C ( see Note 30 ).
Place your imaging dish onto your microscope while illuminating it with 740/750 nm light.
Continuing to bathe the dish in 740/750 nm light, locate the mounted embryos using 488 nm epifluorescence light ( see Note 31 ).
Continuing to bathe the dish in 740/750 nm light and using low-intensity imaging light (488 nm or below, see Note 32 ), find the cells that you want to target.
Allow any background heterodimerization to be reversed under 740/750 nm light and then take an image (or small imaging stack) of your cells (Fig. 5b-ii). Sequentially image lower wavelengths first, followed by any red wavelengths ( see Note 33 ).
![Fig. 5Example subcellular heterodimerization image sequence. (a) Single optogenetically labeled metaphase-stage cell within the zebrafish brain. (b) Image sequence of magnified cell from “A” at a single z-level, demonstrating the steps taken to achieve subcellular heterodimerization. (i) Pard3-EGFP-PIF6 was successfully recruited to the cell membrane under bright-field light. (ii) Pard3-EGFP-PIF6 was sent back into the cytoplasm under 740 nm light (90-s exposure). (iii) An ROI is specified (pink box) and illuminated with low-level 633 nm “binding” laser light while 740 nm “unbinding” light is present globally. This patterned light results in the recruitment of Pard3-EGFP-PIF6 to one half of the dividing cell and consequently to the asymmetric inheritance of this protein into the uppermost cell (dotted lines illustrate the location of the lower cell). This is visualized by periodic imaging under 488 nm light. (c) After a further 24 min of patterned light illumination at a single z-level, a 23 μm z-stack was taken under 488 and 561 nm light to visualize the distribution of Pard3-EGFP-PIF6 (i) and PHYB-MCherry-CAAX (ii) throughout the depth of the cells (note that there is another pair of cells behind the cells of interest, with low-level Pard3-EGFP-PIF6 expression) (adapted and extended from Ref. [17] under the terms of the Creative Commons Attribution License (CC BY): http://creativecommons.org/licenses/by/4.0/) Fig. 5Example subcellular heterodimerization image sequence. (a) Single optogenetically labeled metaphase-stage cell within the zebrafish brain. (b) Image sequence of magnified cell from “A” at a single z-level, demonstrating the steps taken to achieve subcellular heterodimerization. (i) Pard3-EGFP-PIF6 was successfully recruited to the cell membrane under bright-field light. (ii) Pard3-EGFP-PIF6 was sent back into the cytoplasm under 740 nm light (90-s exposure). (iii) An ROI is specified (pink box) and illuminated with low-level 633 nm “binding” laser light while 740 nm “unbinding” light is present globally. This patterned light results in the recruitment of Pard3-EGFP-PIF6 to one half of the dividing cell and consequently to the asymmetric inheritance of this protein into the uppermost cell (dotted lines illustrate the location of the lower cell). This is visualized by periodic imaging under 488 nm light. (c) After a further 24 min of patterned light illumination at a single z-level, a 23 μm z-stack was taken under 488 and 561 nm light to visualize the distribution of Pard3-EGFP-PIF6 (i) and PHYB-MCherry-CAAX (ii) throughout the depth of the cells (note that there is another pair of cells behind the cells of interest, with low-level Pard3-EGFP-PIF6 expression) (adapted and extended from Ref. [17] under the terms of the Creative Commons Attribution License (CC BY): http://creativecommons.org/licenses/by/4.0/)](https://static.yanyin.tech/literature_test/protocol_io_true/protocols.io.bp3jmqkn/370791_2_En_10_Fig5_HTML.png)
Continuing to bathe the dish in 740/750 nm light, define the region of interest (ROI) in which you want heterodimerization to occur ( see Note 34 ).
Set up your imaging program to take a single image (as in step 40), followed by a period of patterned heterodimerization light only (for possible approaches, see Note 29 ), followed by another single image. Repeat these steps for as long as necessary for your experiment (Fig. 5b-iii; see Note 35 ).
Following patterned light illumination you can further image your cells to assess the consequences of the protein recruitment, for example by taking a z-stack (Fig. 5c; see Note 36 ).

