ONT Sequencing IT/Compute Pop!_OS 22.04 Setup
Stephen Douglas Russell
Abstract
The IT requirements for processing MinION data should be carefully reviewed before purchasing a MinION device. You will want to go with a Linux system. System76 is really the primary/best vendor for laptops. Pay careful attention to the GPU. It is probably the most important component for fast processing of the data. Here is is a link to a Facebook thread of some discussion when first considering the specs required.
Setting up all of the programs/dependencies, particularly for utilizing the GPU during the analytical workflows is the next important step. You will want to get all of this in place before you start with the lab workflows, as there are many things that could go wrong or that you will need to work through in order for you to be able to actually begin a run.
Steps
Preparing a new CPU for MinION Sequencing
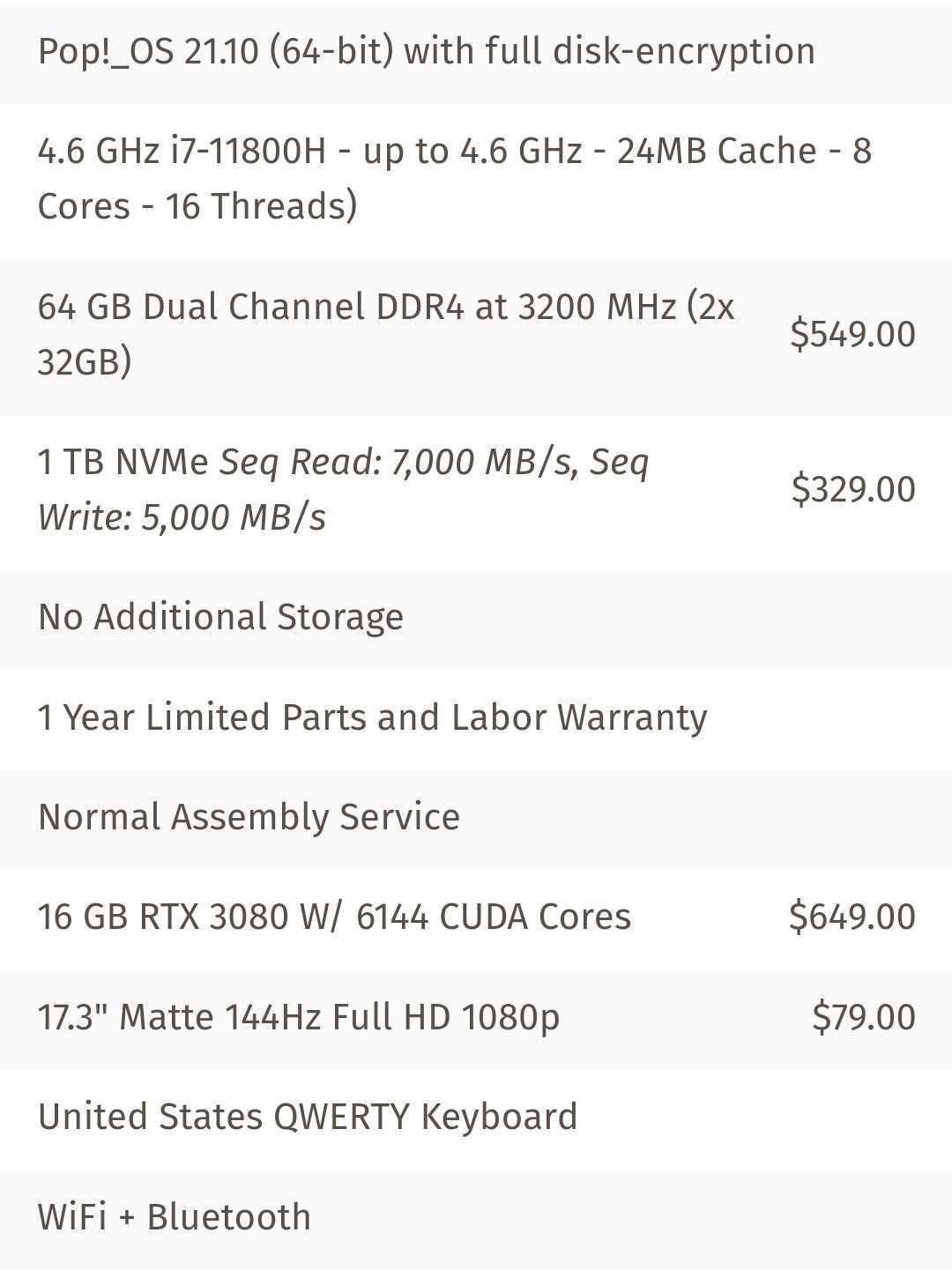
The final setup I went with can be found below. It was expensive (around 4000 for the laptop in early 2022), but should be able to achieve live basecalling for two MinION devices at the same time. Overall specs of my laptop:
Minimum IT requirements for MinION from ONT: minion-it-reqs.pdf
The remainder of this protocol assumes you have completed all of the preliminary setup steps that are common with any new CPU.
Install CUDA toolkit - https://developer.nvidia.com/cuda-toolkit :
wget https://developer.download.nvidia.com/compute/cuda/repos/ubuntu2204/x86_64/cuda-ubuntu2204.pin
sudo mv cuda-ubuntu2204.pin /etc/apt/preferences.d/cuda-repository-pin-600
wget https://developer.download.nvidia.com/compute/cuda/11.7.0/local_installers/cuda-repo-ubuntu2204-11-7-local_11.7.0-515.43.04-1_amd64.deb
sudo dpkg -i cuda-repo-ubuntu2204-11-7-local_11.7.0-515.43.04-1_amd64.deb
sudo cp /var/cuda-repo-ubuntu2204-11-7-local/cuda-*-keyring.gpg /usr/share/keyrings/
sudo apt-get update
sudo apt-get -y install cuda
Install Boost
sudo apt install libboost-all-dev
The process at this link was instrumental to this protocol. It is recreated and simplified here. ORIGINAL PROTOCOL. It was written for Pop!_OS 21.04. The following protocols also work with Pop!_OS 22.04. I would follow the steps at the link rather than here so you get a broader context of the actions you are performing on your system.
#Add ONT Focal Repository (Pop!_OS 22.04)
# update packages list
sudo apt-get update
# check for and install wget if needed
if [ $(dpkg-query -W -f='${Status}' wget 2>/dev/null | grep -c "ok installed") -eq 0 ];
then
sudo apt --yes install wget;
fi
# add the key
wget -O- https://mirror.oxfordnanoportal.com/apt/ont-repo.pub | sudo apt-key add -
# add the focal repo
echo "deb http://mirror.oxfordnanoportal.com/apt focal-stable non-free" | sudo tee /etc/apt/sources.list.d/nanoporetech.sources.list
#sudo apt update (Pop!_OS 22.04)
sudo apt update
#Check for access to ONT files (Pop!_OS 22.04)
apt policy minknow-core-minion-nc
Add the Focal repos:
#Create a new file and edit in nano
sudo nano /etc/apt/sources.list.d/system-focal.sources
```Copy and paste the following into your file:
```X-Repolib-Name: Pop_OS System Sources<br/>Enabled: yes<br/>Types: deb deb-src<br/>URIs: http://us.archive.ubuntu.com/ubuntu/<br/>Suites: focal focal-security focal-updates focal-backports<br/>Components: main restricted universe multiverse<br/>X-Repolib-Default-Mirror: http://us.archive.ubuntu.com/ubuntu/
cat system-focal.sources
Pin the Focal repos. Start by creating another new file with nano:
sudo nano /etc/apt/preferences.d/focal-default-settings
Copy and paste the following into your file:
Check that is was created correctly
cat focal-default-settings
sudo apt update
Install MinKNOW and required packages
sudo apt install \
minknow-core-minion-nc \
ont-kingfisher-ui-minion \
ont-bream4-minion \
ont-configuration-customer-minion \
ont-jwt-auth \
ont-vbz-hdf-plugin
Install ONT Guppy
sudo apt install ont-guppy
which guppy_basecaller
guppy_basecaller --version
Setup the MinKnow service
sudo /opt/ont/minknow/bin/config_editor --conf application \
--filename /opt/ont/minknow/conf/app_conf \
--set guppy.server_executable="/opt/ont/guppy/bin/guppy_basecall_server" \
--set guppy.client_executable="/opt/ont/guppy/bin/guppy_basecall_client" \
--set guppy.gpu_calling=1 \
--set guppy.num_threads=16 \
--set guppy.ipc_threads=2
systemctl restart minknow.service
systemctl status minknow.service
sudo nano /lib/systemd/system/guppyd.service
```Copy the following to your new file:
```[Unit]<br/>Description=Service to manage the guppy basecall server.<br/>Documentation=https://community.nanoporetech.com/protocols/Guppy-protocol/v/GPB_2003_v1_revQ_14Dec2018<br/><br/>[Service]<br/>Type=simple<br/>ExecStart=/opt/ont/guppy/bin/guppy_basecall_server --log_path /var/log/guppy --config dna_r9.4.1_450bps_fast.cfg --port 5555 -x cuda:all <br/>Restart=always<br/>User=root<br/>MemoryLimit=8G<br/>MemoryHigh=8G<br/>CPUQuota=200%<br/><br/>[Install]<br/>Alias=guppyd.service<br/>WantedBy=multi-user.target
Check the file:
cat /lib/systemd/system/guppyd.service
systemctl enable guppyd.service
systemctl restart guppyd.service
MinKNOW GUI should now be available in your programs. Validate that it opens correctly.
Changes to MinKnow file permissions at the bottom here:
https://gringer.gitlab.io/presentation-notes/2021/10/08/gpu-calling-in-minknow/
"For my computer, there’s an issue with MinKNOW not being able to access or create files. As a “nuclear” option, Miles Benton suggested changing the user and group for the minknow service to root"
sudo service minknow stop
sudo perl -i -pe 's/(User|Group)=minknow/$1=root/' /lib/systemd/system/minknow.service
sudo systemctl daemon-reload
sudo service minknow start
Per this document: https://denbi-nanopore-training-course.readthedocs.io/en/latest/read_qc/MinionQC.html
Install R: https://cran.r-project.org/
Install MinionQC: https://github.com/roblanf/minion_qc
Install R:
# update indices
sudo apt update -qq
# install two helper packages we need
sudo apt install --no-install-recommends software-properties-common dirmngr
# add the signing key (by Michael Rutter) for these repos
# To verify key, run gpg --show-keys /etc/apt/trusted.gpg.d/cran_ubuntu_key.asc
# Fingerprint: E298A3A825C0D65DFD57CBB651716619E084DAB9
wget -qO- https://cloud.r-project.org/bin/linux/ubuntu/marutter_pubkey.asc | sudo tee -a /etc/apt/trusted.gpg.d/cran_ubuntu_key.asc
# add the R 4.0 repo from CRAN -- adjust 'focal' to 'groovy' or 'bionic' as needed
sudo add-apt-repository "deb https://cloud.r-project.org/bin/linux/ubuntu $(lsb_release -cs)-cran40/"
sudo apt install --no-install-recommends r-base
install.packages(c("data.table",
"futile.logger",
"ggplot2",
"optparse",
"plyr",
"readr",
"reshape2",
"scales",
"viridis",
"yaml"))
Install Bioconductor:
In an R command window:
if (!require("BiocManager", quietly = TRUE))
install.packages("BiocManager")
BiocManager::install(version = "3.15")
Install Anaconda:
cd /tmp
curl https://repo.anaconda.com/archive/Anaconda3-2022.05-Linux-x86_64.sh --output anaconda.sh
sha256sum anaconda.sh
bash anaconda.sh
source ~/.bashrc
conda list
conda search "^python$"
conda create --name my_env python=3
conda activate my_env
python --version
conda install --name my_env35 numpy
Install NGSpeciesID: https://github.com/ksahlin/NGSpeciesID
conda create -n NGSpeciesID python=3.6 pip
conda activate NGSpeciesID
conda install --yes -c conda-forge -c bioconda medaka==0.11.5 openblas==0.3.3 spoa racon minimap2
pip install NGSpeciesID
conda activate NGSpeciesID
mkdir test_ngspeciesID
cd test_ngspeciesID
curl -LO https://raw.githubusercontent.com/ksahlin/NGSpeciesID/master/test/sample_h1.fastq
NGSpeciesID --ont --fastq sample_h1.fastq --outfolder ./sample_h1 --consensus --medaka
You should now be ready to begin sequencing runs.
Updating MinKnow & Installing Dorado
Since this protocol was first written, MinKnow needed an update for the 10.4.1 Flongle cells and then again to bring in Dorado basecaclling. The steps to perform the update can be found below:
sudo apt install ont-python=3.10.7-9
sudo apt install ont-bream4-minion=7.8.2-1~focal
sudo apt install ont-configuration-customer-minion=5.8.6-1~focal
sudo apt install ont-kingfisher-ui-minion=5.8.13-1~focal
sudo apt install minknow-core-minion-nc=5.8.3
sudo apt update
sudo apt install wget
wget -O- https://cdn.oxfordnanoportal.com/apt/ont-repo.pub | sudo apt-key add -
echo "deb http://cdn.oxfordnanoportal.com/apt focal-stable non-free" | sudo tee /etc/apt/sources.list.d/nanoporetech.sources.list
sudo apt update
sudo apt install ont-standalone-minknow-gpu-release
Carryover code from last update:
sudo service minknow stop
sudo perl -i -pe 's/(User|Group)=minknow/$1=root/' /lib/systemd/system/minknow.service
sudo systemctl daemon-reload
sudo service minknow start
sudo apt install cuda
conda install -c bioconda seqkit
python -m venv venv --prompt duplex
. venv/bin/activate
pip install duplex_tools
These dependency versions will likely keep changing with time.
Installing Dorado:
sudo -i
sudo apt-get update && apt-get install -y --no-install-recommends \
curl \
git \
ca-certificates \
build-essential \
nvidia-cuda-toolkit \
libhdf5-dev \
libssl-dev \
libzstd-dev \
cmake \
autoconf \
automake
git clone https://github.com/nanoporetech/dorado.git dorado
cd dorado
cmake -S . -B cmake-build
cmake --build cmake-build --config Release -j
ctest --test-dir cmake-build
pip install pre-commit
pre-commit install

