Non-invasive Detection Method for Bonamia ostreae in Ostrea edulis using the Franklin qPCR machine
Lavanya M Vythalingam, Tim Regan, Tim Bean
Abstract
This is a comprehensive protocol for using the Franklin qPCR (Biomeme Inc.) machine to detect the presence of Bonamia ostreae parasite in European flat oyster ( Ostrea edulis ). Previous methods relied on sacrificially dissecting animals to perform histology. While sensitive, these methods were destructive and untested animals carrying Bonamia ostreae are not detected. Here, we present a non-invasive alternative method to detect Bonamia ostreae from Ostrea edulis pseudofaeces/faeces using a portable qPCR machine.
Steps
Sample Collection
Collect oysters and wash to remove any excess mud/sediment on their shells.
Separate collected animals and cover with seawater (approx. 1L for every 200g of live animal).
Collect the sediment remaining in the bottom of each bucket (faeces and pseudofaeces) using a Pasteur pipette into a 1.5mL Eppendorf tube.
OPTIONAL: Store samples collected at -20°C until further use.
Disinfect all equipment with working strength bleach according to biosecurity SOPs.
DNA Extraction
Extract DNA from the sediment samples by physical beating in 2mL Lysing matrix A tubes and filter through a 1 uM barb column filter.
Use a syringe with a 1 uM pre-filter to remove the supernatant without debris.
Make two holes in the first M1 chamber and transfer sample into the Biomeme M1 cartridge.
Process filtered sediment through the Biomeme M1 extraction cartridge (see Figure 2), according to the manufacturer’s protocol (refer to link below) .
M1 Sample Prep Cartridge Techniques on Vimeo

On-site Franklin qPCR assay
Store DNA at -4°C until further use.
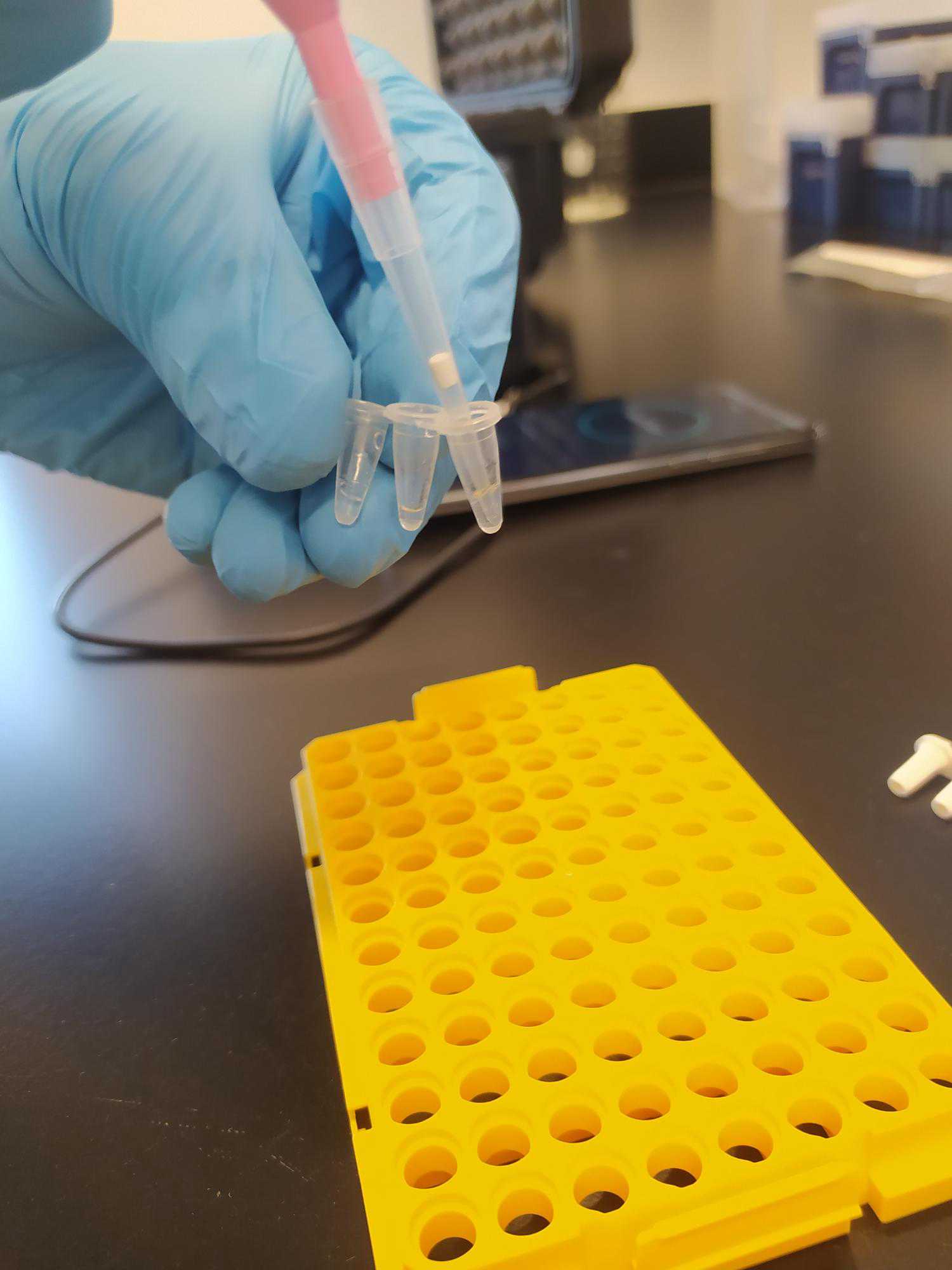
Transfer 20µL of extracted DNA to Biomeme Go-Strip assay tube (see Figure 3).
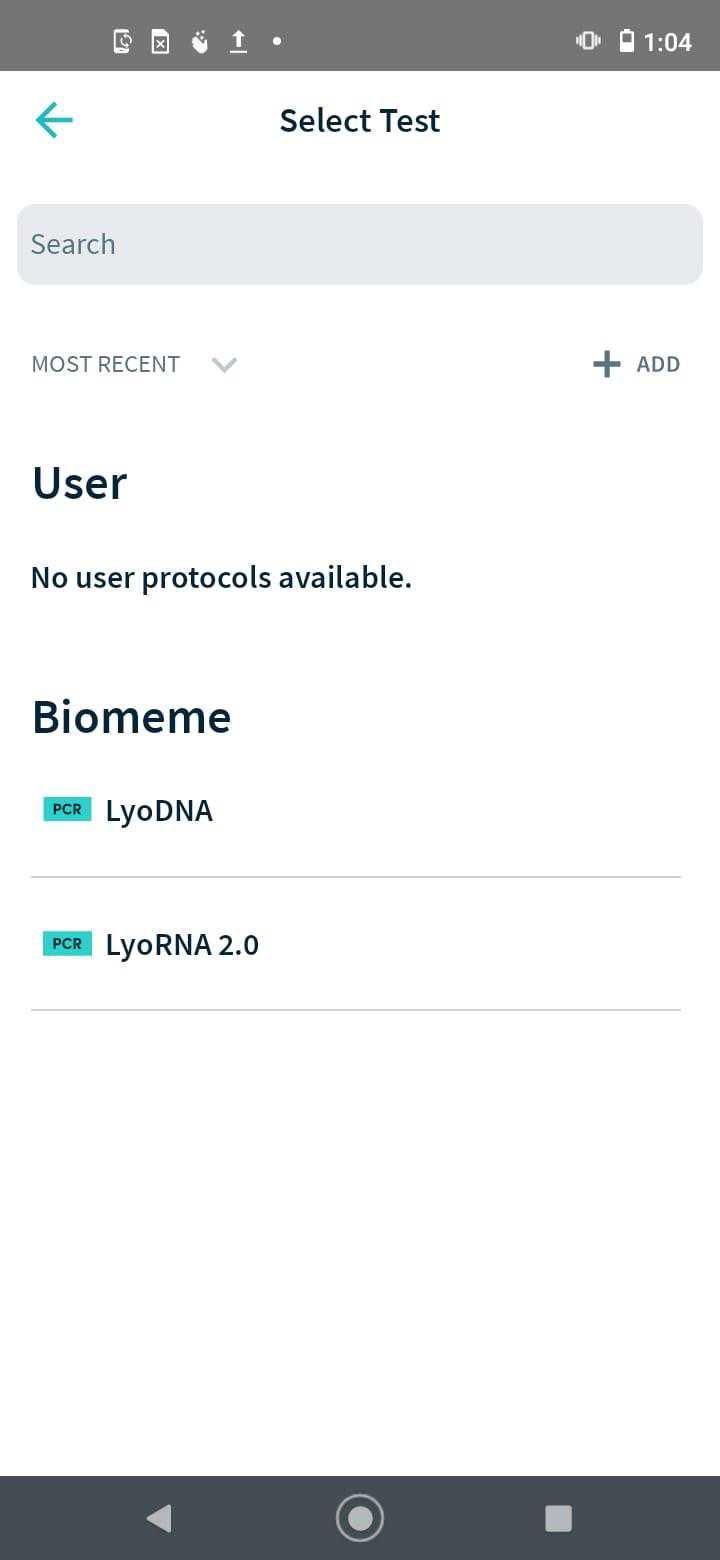
Run a qPCR assay using the Franklin qPCR machine against the three probe-based assays. Primers and probes used in this assay is given below (Table 1).
The Franklin qPCR protocol consisted of 0h 1m 0s minute heat activation and 45 cycles of 95°C for 1 second and 60°C for 0h 0m 20s.
Use the Biomeme app on the phone to analyse the qPCR results to determine the presence of Bonamia ostreae. Refer to Figure 6 for an example of positive Bonamia ostreae presence .