Nanopore amplicon sequencing
Yoshiyuki Matsuo
Abstract
The two-step PCR method allows us to perform nanopore amplicon sequencing with a user-defined inner primer set combined with barcoded outer primers provided by Oxford Nanopore Technologies, taking advantage of rapid adapter attachment chemistry. This method can be applied to a wide range of sequence-based analyses, including microbiome profiling and the identification of genetic variations in targeted loci.
Steps
Workflow
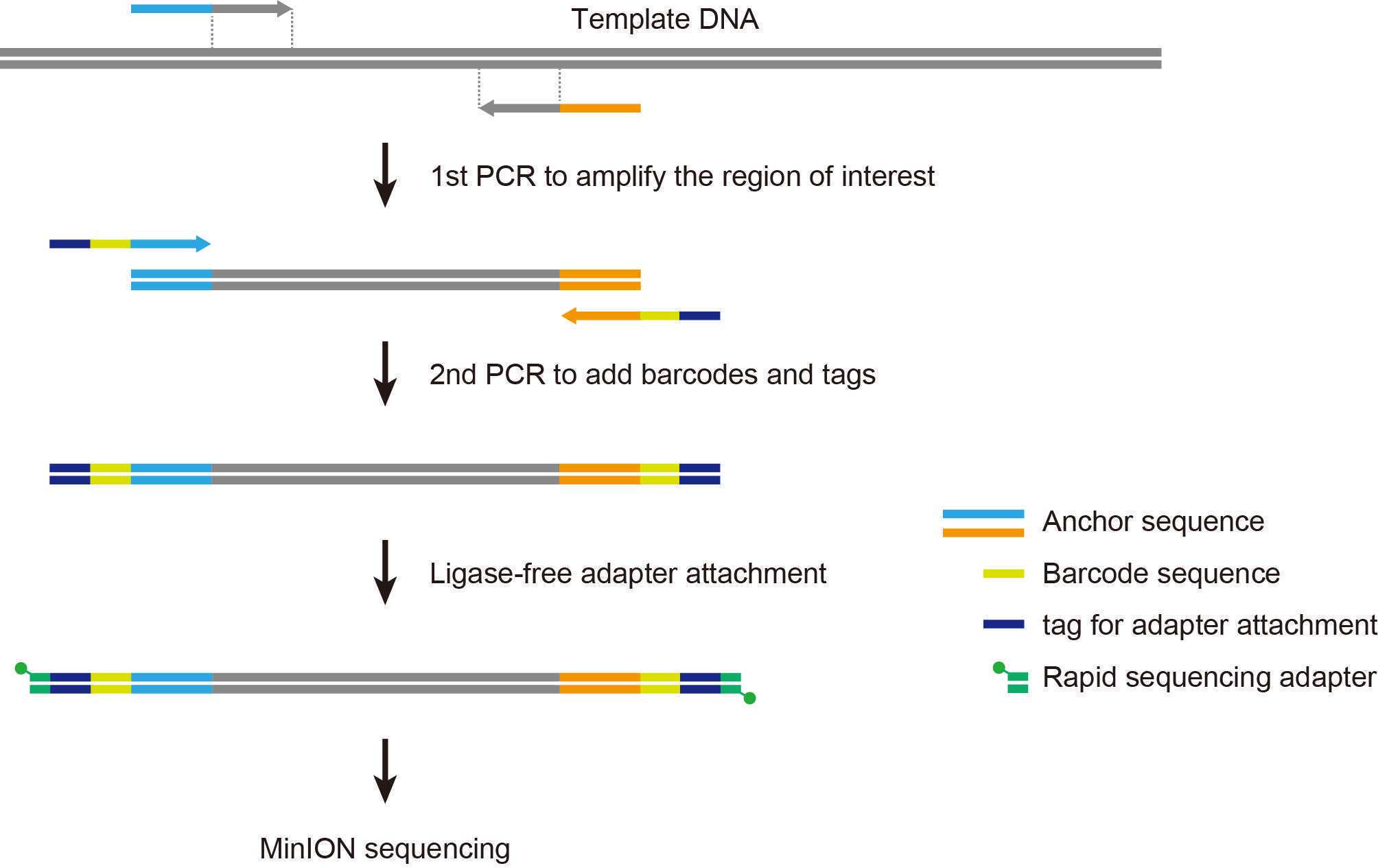
1st PCR with inner primers
Prepare the PCR master mix.
| A | B | C |
|---|---|---|
| Component | Volume | Final conc. |
| Template DNA | x µL | |
| 10 µM FW/RV primer mix | 0.5 µL | 0.2 µM each |
| 2X KAPA2G Robust HS ReadyMix | 12.5 µL | 1X |
| Water | 12 - x µL | |
| Total | 25 µL |
Inner primers (user-supplied)
| A | B |
|---|---|
| Primer | Sequence |
| Forward (FW) | 5'-TTTCTGTTGGTGCTGATATTGC - target-specific sequence -3' |
| Reverse (RV) | 5'-ACTTGCCTGTCGCTCTATCTTC - target-specific sequence -3' |
The 5' anchor sequences serve as priming sites for barcoded outer primers used in the 2nd PCR.
Perform PCR.
| A | B | C | D |
|---|---|---|---|
| Step | Temperature | Time | Cycles |
| Initial denaturation | 95°C | 3 min | 1 |
| Denaturation | 95°C | 15 sec | 25-35 |
| Annealing | 55°C | 15 sec | |
| Extension | 72°C | 30 sec | |
| Hold | 4°C | ∞ | 1 |
The above is an example for amplifying the near-full length (V1–V9) sequence of bacterial 16S rRNA genes (approximately 1,500 bp).
Equipment
| Value | Label |
|---|---|
| Veriti 96-Well Thermal Cycler | NAME |
| Applied Biosystems | BRAND |
| 4375786 | SKU |
| https://www.thermofisher.com/us/en/home.html | LINK |
Analyze 2µL of the PCR products by gel electrophoresis to verify successful amplification.
Equipment
| Value | Label |
|---|---|
| E-Gel Power Snap Electrophoresis Device | NAME |
| Thermo Fisher Scientific | BRAND |
| G8100 | SKU |
| https://www.thermofisher.com/us/en/home.html | LINK |
Equipment
| Value | Label |
|---|---|
| E-Gel Power Snap Camera | NAME |
| Thermo Fisher Scientific | BRAND |
| G8200 | SKU |
| https://www.thermofisher.com/us/en/home.html | LINK |
2nd PCR with barcoded outer primers
Prepare the PCR master mix.
| A | B |
|---|---|
| Component | Volume |
| 1st PCR products | 1.0 µL |
| BP01–12 | 0.5 µL |
| 2X KAPA2G Robust HS ReadyMix | 12.5 µL |
| Water | 11 µL |
| Total | 25 µL |
BP01–12: barcoded outer primers supplied in the PCR Barcoding Kit.
Perform PCR.
| A | B | C | D |
|---|---|---|---|
| Step | Temperature | Time | Cycles |
| Initial denaturation | 95°C | 3 min | 1 |
| Denaturation | 95°C | 15 sec | 8–10 |
| Annealing | 62°C | 15 sec | |
| Extension | 72°C | 30 sec | |
| Hold | 4°C | ∞ | 1 |
The above is an example for barcoding bacterial 16S rRNA gene amplicons (approximately 1600 bp).
Equipment
| Value | Label |
|---|---|
| Veriti 96-Well Thermal Cycler | NAME |
| Applied Biosystems | BRAND |
| 4375786 | SKU |
| https://www.thermofisher.com/us/en/home.html | LINK |
Analyze 1µL of the PCR products by gel electrophoresis.
Equipment
| Value | Label |
|---|---|
| E-Gel Power Snap Electrophoresis Device | NAME |
| Thermo Fisher Scientific | BRAND |
| G8100 | SKU |
| https://www.thermofisher.com/us/en/home.html | LINK |
Equipment
| Value | Label |
|---|---|
| E-Gel Power Snap Camera | NAME |
| Thermo Fisher Scientific | BRAND |
| G8200 | SKU |
| https://www.thermofisher.com/us/en/home.html | LINK |
PCR cleanup
Resuspend the AMPure XP beads by vortexing.
Add AMPure XP beads to the sample and mix by pipetting.
| A | B |
|---|---|
| Component | Volume |
| 2nd PCR products | 24 µL |
| AMPure XP | 12 µL |
Incubate at Room temperature for 0h 5m 0s.
Place the tube on a magnetic rack for 0h 2m 0s.
Equipment
| Value | Label |
|---|---|
| NGS MagnaStand v.3 8Ch | NAME |
| Magnetic rack (0.2 mL tube) | TYPE |
| FastGene | BRAND |
| FG-SSMAG3 | SKU |
Pipette off the supernatant.
Wash the beads with 70% ethanol as follows (1/2).
Keeping on the magnetic rack, add 200µL of 70% ethanol without disturbing the bead pellet.
Discard the supernatant.
Wash the beads with 70% ethanol as follows (2/2).
Keeping on the magnetic rack, add 200µL of 70% ethanol without disturbing the bead pellet.
Discard the supernatant.
Spin down and place the tube back in the magnetic rack.
Pipette off any residual ethanol.
Remove the tube from the magnetic rack and resuspend the beads in 10µL of TN buffer.
Incubate at Room temperature for 0h 2m 0s.
Place the tube on a magnetic rack for 0h 2m 0s .
Transfer the eluate to a new tube.
[Optional] Analyze 1µL of the purified sample by gel electrophoresis to confirm the recovery.
DNA quantification
Warm QuantiFluor ONE dsDNA dye to Room temperature .
Add 1µL of eluted sample to 200µL of QuantiFluor ONE dsDNA dye in 0.5 mL tube.
Mix thoroughly by vortexing.
Incubate at Room temperature for 0h 5m 0s, protected from light.
Measure fluorescence using the Quantus Fluorometer to quantify DNA concentration.
Equipment
| Value | Label |
|---|---|
| Quantus Fluorometer | NAME |
| Promega | BRAND |
| E6150 | SKU |
| http://www.promega.com | LINK |
Sequencing library preparation
Pool all barcoded amplicons to a total of 50-100fmoles in 10µL of TN buffer.
| A | B | C |
|---|---|---|
| Component | Volume | DNA |
| Sample #01 (25 ng/µL) | 1.0 µL | 25 ng |
| Sample #02 (25 ng/µL) | 1.0 µL | 25 ng |
| Sample #03 (25 ng/µL) | 1.0 µL | 25 ng |
| Sample #04 (25 ng/µL) | 1.0 µL | 25 ng |
| TN buffer | 6.0 µL | - |
| Total | 10 µL | 100 ng |
In the above example, four barcoded 16S rRNA gene amplicons (~1600 bp) are pooled together in equal proportions.
Add 1µL of Rapid Adapter (RAP) and mix gently by pipetting.
Incubate at Room temperature for 0h 5m 0s.
Store the library On ice until ready to load.
Flow cell check
Open the MinION lid and insert the flow cell under the clip.
Equipment
| Value | Label |
|---|---|
| MinION Mk1C | NAME |
| Oxford Nanopore Technologies | BRAND |
| M1CBasicSP | SKU |
| https://nanoporetech.com/ | LINK |
Perform flow cell check.
Check the number of active pores available for the experiment.
Sample loading
Prepare flow cell priming mix and vortex thoroughly.
| A | B |
|---|---|
| Component | Volume |
| Flush Tether (FLT) | 30 µL |
| Flush Buffer (FB) | 1.17 mL |
| Total | 1.2 mL |
FLT and FB are supplied in the Flow Cell Priming Kit. FB is provided in tubes, pre-aliquoted with 1.17 mL.
Open the priming port cover of the flow cell.
Remove air bubbles under the cover as follows (if any).
Set the volume of P1000 micropipette to 200 µL.
Insert the tip into the priming port.
Turn the wheel of the pipette slowly to increase the volume and draw back 20-30µL of the buffer.
Load 800µL of the priming mix (from Step 33) into the flow cell via the priming port.
Wait for 0h 5m 0s.
Prepare the sequencing library for loading.
| A | B |
|---|---|
| Component | Volume |
| Library (from Step 29) | 11 µl |
| Water | 4.5 µl |
| Sequencing Buffer (SQB) | 34 µl |
| Loading Beads (LB) | 25.5 µl |
| Total | 75 µl |
SQB and LB are supplied in the PCR Barcoding Kit.
Lift the SpotON sample port cover of the flow cell.
Load 200µL of the priming mix (from Step 33) into the flow cell via the priming port (caution: not the SpotON sample port).
Gently mix the sequencing library (75µL, prepared in Step 38) by pipetting just prior to loading.
Load the library into the flow cell via the SpotON sample port in a dropwise fashion.
Replace the SpotON sample port cover and close the priming port.
Nanopore sequencing
Start sequencing run.
| A | B |
|---|---|
| Parameter | Setting |
| Flow cell type | FLO-MIN106 (R9.4.1) |
| Kit | PCR Barcoding Kit SQK-PBK004 |
| Basecalling | On |
| Basecalling configuration | Fast basecalling |
| Barcoding | On |
| Trim barcodes | On |
| Barcode both ends | Off |
| Mid-read barcode filtering | On |
| Q score filtering | 8 (default value) |
Typical examples of run parameters with real-time basecalling on the MinION Mk1C.
Flushing a flow cell
Stop sequencing run.
Prepare flow cell wash mix and gently mix by pipetting.
| A | B |
|---|---|
| Component | Volume |
| Wash Mix (WMX) | 2 µL |
| Wash Diluent (DIL) | 398 µL |
| Total | 400 µL |
WMX and DIL are supplied in the Flow Cell Wash Kit. WMX contains DNase I.
Remove fluid in the waste channel via the waste port.
Open the priming port cover of the flow cell.
[Optional] If necessary, remove air bubbles under the cover by following the procedure in Step 35.
Load 400µL of the wash mix (from Step 46) into the flow cell via the priming port.
Close the priming port.
Wait for 1h 0m 0s to digest remaining DNA on the flow cell.
Remove fluid in the waste channel via the waste port.
Open the priming port cover of the flow cell.
[Optional] If necessary, remove air bubbles under the cover by following the procedure in Step 35.
Load 500µL of Storage Buffer (S) into the flow cell via the priming port.
Close the priming port.
Remove fluid in the waste channel via the waste port.
Store the flow cell at 4°C for subsequent use.

