Multiplex Real-Time PCR for genotyping Glutathione S-Transferase deletion polymorphisms
Rayana Pereira Dantas de Oliveira, Kamilla de Faria Santos, Wandelisa Cançado Flores Menezes, Rodrigo da Silva Santos, Angela Adamski da Silva Reis
Abstract
Glutathione S-transferases (GSTs) are enzymes that act in the conjugation of reactive metabolites to the reduced glutathione. The GSTM1 and GSTT1 deletion polymorphisms promote a detriment in the metabolic detoxification process of xenobiotics, due to present a non-functional null allele, causing oxidative stress, may be related to the pathophysiology of various diseases. Genetic association case-control studies usually determine whether a statistical association exists between the disease trait and the genetic marker, such as GST deletion polymorphism. Therefore, techniques to genotyping these polymorphisms through multiplex PCR are highly desired and of wider interest. This method allows for the detection of genotypes for genetic association studies in molecular pathology and it is more cost-effective than hybridization probes.

Before start
Pay attention to all necessary precautions to perform a PCR assay.
Steps
Description of protocol
Glutathione S-transferases (GSTs) are members of a multigene family of phase II enzymes, which are responsible for coding isoenzymes that protect the organism against oxidative damage [1]. The expression of GSTs enzymes is mainly affected by the genetic polymorphisms. Polymorphic variants in the GSTM1 (GSTM10) and GSTT1 (GSTT10) genes produce either a functional proteins, i.e. non null phenotypes (non-deletion alleles or heterozygous deletion) or result in the complete absence of enzymes, i.e. null phenotypes (homozygous deletion or null genotype) [2].
The homozygous deletions in GSTM1 and GSTT1 genes are associated with reduced detoxification function and increased susceptibility to oxidative damage, due the absence of the enzymatic activity of these two isoforms. Thus, the oxidative stress, as a potential cause of cellular dysfunction, may be related to the pathophysiology of various diseases [3].
Current methodologies used to genotyping GST deletion polymorphisms are largely based on “conventional Polymerase Chain Reaction (PCR)". Most of the studies perform an initial endpoint PCR amplification of the region of interest. Subsequently, an aliquot of the post-PCR product is used to further confirmatory by simple electrophoresis [4,5]. However, this technique is laborious to identify the genotype null. Separation of PCR products by gel electrophoresis no presents the complication of discriminating between wild-type and null genotypes and housekeep gene to discriminate the absence of amplification. The multiplex Real-Time-PCR by SYBR Green® is a technique more easy, fast to perform and more reproducible and less expensive than other methods. In this document, we share our protocol for amplification to these deletion polymorphisms.
Abdel-Rahmana et al. [6] and Marin et al. [7] proposed the protocols for multiplex genotyping to GST deletions polymorphism. This last was proposed for real-time PCR. Thus, we adapted these two protocols to multiplex real-time PCR using another gene as internal amplification control to exclude false negatives, were simultaneously amplified, and other alterations. Particularly, this protocol is promising because it is a simple and more economical method than hybridization probes.
Samples collection and DNA extraction
Peripheral blood samples from diabetic patients and health control group were collected in tubes containing heparin and stored at -80°C to preserve the characteristics of nucleic acid for genotyping. The DNA extraction was performed using the PureLink ™ Genomic DNA Mini Kit (Thermofisher, USA) following the manufacturer’'s suggested protocol. All DNA samples were previously quantified in Nanodrop ™ ND-1000 (Thermofisher, USA) for genotyping.
Primer design
The set of primers were adapted from Pinheiro et al. [3], Abdel-Rahman et al. [6] and Marín et al. [7], and are described in Table 1.
| A | B | C | D |
|---|---|---|---|
| Gene | Sequence (5’- 3`) | PCR product size (bp) | Reference |
| GSTM1 | GAA CTC CCT GAA AAG CTA AAG C | 215 | [6;7] |
| GSTT1 | TTC CTT ACT GGT CCT CAC ATC TC | 257 | [7] |
| RH92600 | GCA ATT CCG CAT TTA ATT CAT GG | 135 | [3] |
Table 1. Primers Sequences
These primers were evaluated, the in silico validation was performed with the BLAST tool and the Gene Runner software to verify their efficiency and specificity, which is essential for accurate quantification in qPCR. Each primer set works best under different concentration conditions. Therefore, for this protocol, the concentrations of the primers were defined as ideal when we obtained maximum specific amplification compared to the primer dimers in an experiment using positive control versus negative control.
Multiplex amplification by real-time PCR
In the performed protocol, approximately 10ng of DNA from previously quantified and diluted samples were as added, to the reaction mix. For the final volume of 25 μL, 0.3 μM of each primer forward (F) and reverse (R) of GSTT1, 0.4μM F and R of GSTM1 and 0.5 μM of each primer RH92600 were added. In addition, 1.25 mM MgCl2 and 1x of SYBR Green qPCR Master Mix (Sso Advanced™ Universal SYBR® Green Supermix- Bio-Rad, USA), a fluorescent dye that binds to the double strand of DNA (dsDNA), were added.
The qPCR was performed in the IQ5 thermocycler (Bio-Rad®, USA) with simultaneous amplification of the primers, with the co-amplification of the RH92600 gene, a microsatellite region, used as an endogenous control of the reaction to exclude false negative results. The cycling conditions followed the following steps: initial denaturation at 95 °C for 10 min, followed by 33 cycles by denaturation at 95 °C for 10 s; annealing at 60 °C for 20 s and extension for 25 s at 72 °C.
Melting curve analysis
As SYBR® Green nests to every dsDNA, an increase in fluorescence intensity is produced proportional to the number of amplicons [8]. Therefore, each amplified sequence is characterized by the melting temperature (Tm) which varies depending on the length and sequence bases of the amplified product. Tm represents the temperature at which half of the target DNA is duplicated and the other half is dissociated, generating a fluorescence decay curve called melting curve [7].The programming performed for the melting curve was: 95 °C for 10 s, 65 °C for 1 min and increase to 95 °C, with 5 acquisitions per °C. The Tm analysis was performed using the Bio-Rad® IQ5 software.
The generation and comparison of the melting curves in the amplification process is a method that increases the specificity of the multiplex qPCR technique with SYBR® Green. Each amplicon has a characteristic Tm that differentiates them from unexpected double-stranded DNA amplification (primer dimer, hairpin), as it denatures at lower temperatures [8]. Therefore, we identified the following Tm for the analyzed genes: RH92600 (83°C), GSTM1 (87°C) and GSTT1 (89°C).
Each amplification group was performed with a negative control without a DNA sample and a positive control with a previously known genotype sample. The discrimination of the null and present genotypes by analysis of the melting curves generated after the amplification reactions (Figure 1).
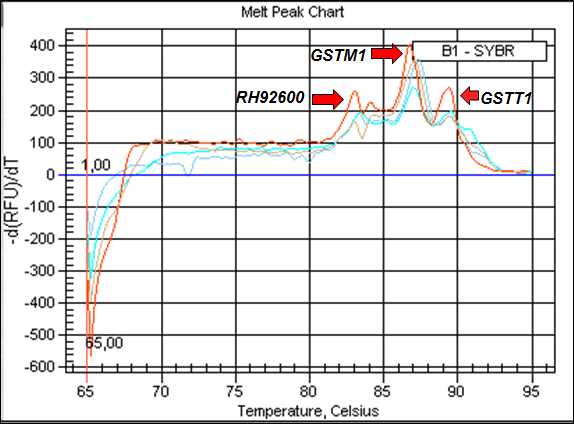
Figure 1 describes the melt curve and PCR products amplified from the 3 genes, RH92600, GSTM1 and GSTT1, respectively. For absence of peaks corresponding to GSTM1 and/or GSTT1, in the presence of endogenous control (RH92600), allows the identification of the respective GSTM1 and ⁄or GSTT1 null genotypes. For each DNA sample, we did a biological replica to guarantee the results of the genotypes evaluated.

