Modular generation of cortical, striatal and ventral midbrain progenitor cells
Carles Calatayud, Esther Muñoz-Pedrazo, Sandra Fernández-Gallego, Patrik Verstreken
A9 dopamine neurons
cortical neurons
striatal neurons
medium spiny neurons
hiPSC
differentiation
lateral ganglionic eminence
Abstract
The present protocol describes the modular generation of cortical, striatal, and ventral midbrain human neural progenitor cells from human-induced pluripotent stem cells (hiPSC). The different elements of the differentiation protocols have been optimized so that they only differ in the patterning factors (protein ligands or small molecules) that are required for each lineage.
Steps
Preparation of the hiPSC for starting the neuralization
Notes before starting:
- We maintain hPSC in mTESR-Plus or StemFlex medium on Matrigel-coated plates and split the cells as clumps using RELESR or 0.5 mM EDTA. Accutase is used when cells need to be counted such as in the initial step of the protocol. Or for the first passages of the NPCs (breaking the monolayer). Accutase needs to be 1) fresh and 2) washed by dilution and centrifugation. It is advisable to prolong accutase treatment (7 to 10 minutes) rather than singularize cells by repeated pipetting.
- It is important to coat the plates with fresh Matrigel for the neuralization step (days 0 to 11). Otherwise, the borders of the monolayer may detach during the course of the neuralization
- In order to have enough cells for the initial seeding, we generally grow the cells in 60mm or 100mm plates. A confluent 100mm contains enough cells to seed an entire 6 well plate.
Dual-SMAD inhibition and regional patterning
Day 0:
- Pretreat the PSC with RI for 2h to O/N.
- Remove the medium (this medium can be saved for diluting the accutase).
- Add 3 mL of pre-warmed accutase to a 100mm plate (alternatively 1.5 mL to a 6mm). Incubate the cells with the accutase for 5-10' in the incubator at 37oC. Before cells start to detach, remove the accutase. Pipette medium (fresh or the medium saved before) on top of the cells vigorously in order to detach them from the plate. It is very important not to pipette medium with cells suspended in it as this would harm the cells.
- Count the cell suspension, and Seed 400k cells per cm2 in the medium formulation indicated in the graph below in the presence of RI. Days 1-6: change the medium on day 1 to remove RI. Refresh medium every other day (3 mL per 6wp well).
Days 6-11: change medium every day (3 mL per 6wp well). Floor plate NPCs may require 4-5 mL of medium.
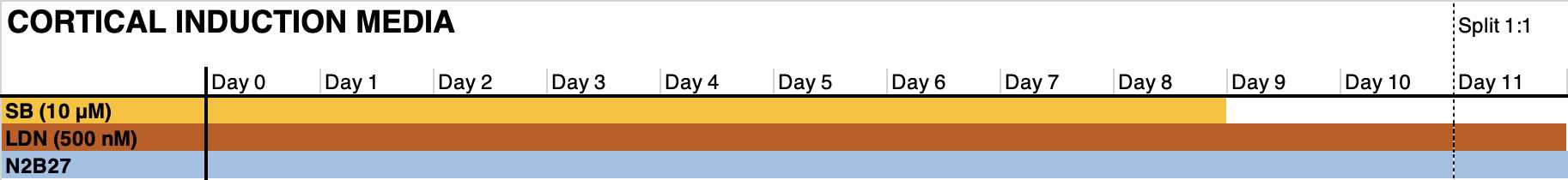
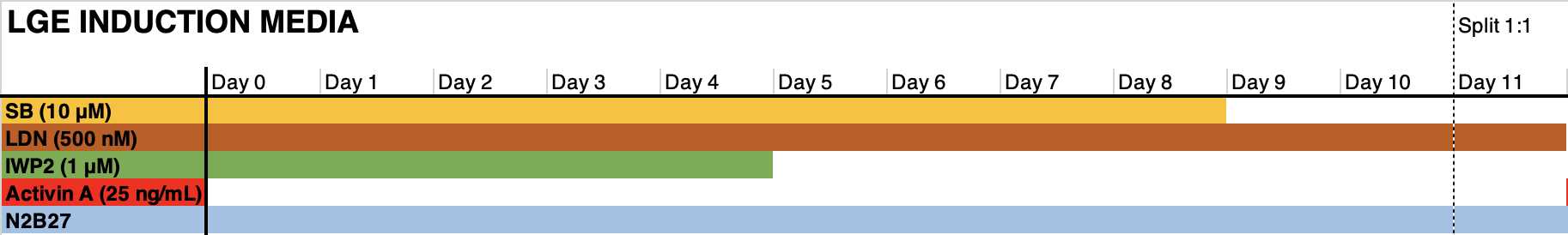
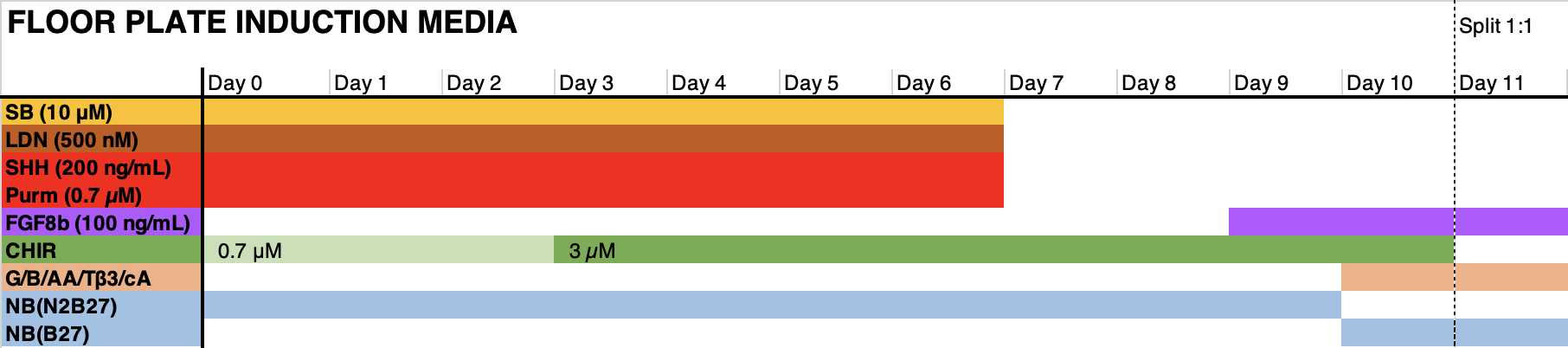
Expansion and subsequent cryopreservation of neural progenitor cells.
Day 11: reseed cells 1:1 in a matrigel-coated plate using accutase. This cell passaging step facilitates the homogenization of the differentiating cells and the elimination of dead cells trapped in the monolayer. Pre- and post- incubate cells with RI. Do not seed less than 800.000 cells/cm2
Day 12: refresh medium to remove RI from the medium.
Day 14: refresh medium.
Day 16: preincubate with RI from 2h to O/N. Split cells 1:2 if possible using accutase; do not seed less than 800.000 cells/cm2. Reseed in medium supplemented with RI.
--> Optional: floor Plate progenitors can be QCed at this stage since they already express relevant region-specific markers
Day 17: refresh medium to remove RI to the medium.
Day 19: refresh medium
Day 20: preincubate with RI from 2h to O/N. freeze cells in freezing medium. Reserve 1:6 of the cells for seeding in 2-3x 24wp wells and for RNA analysis.
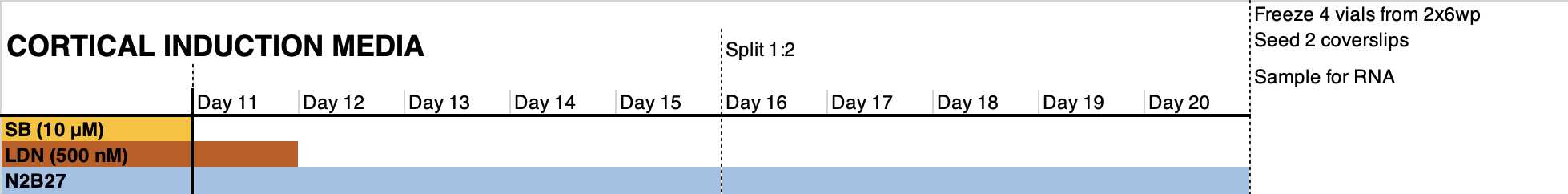
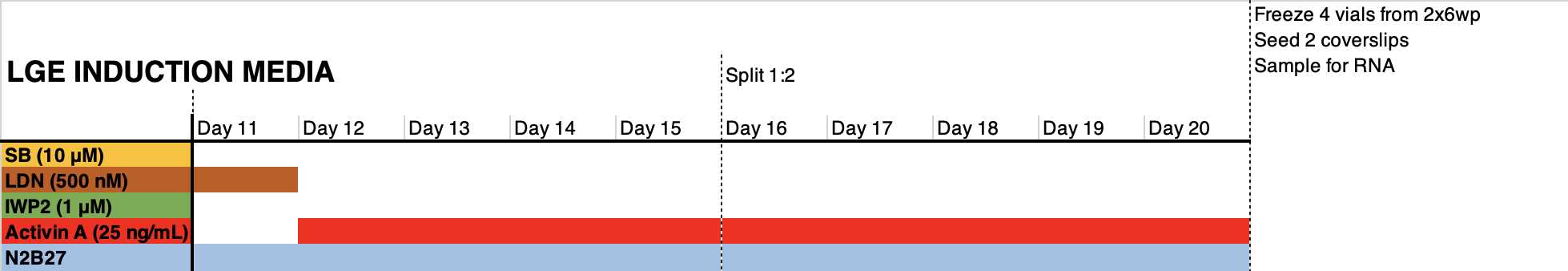
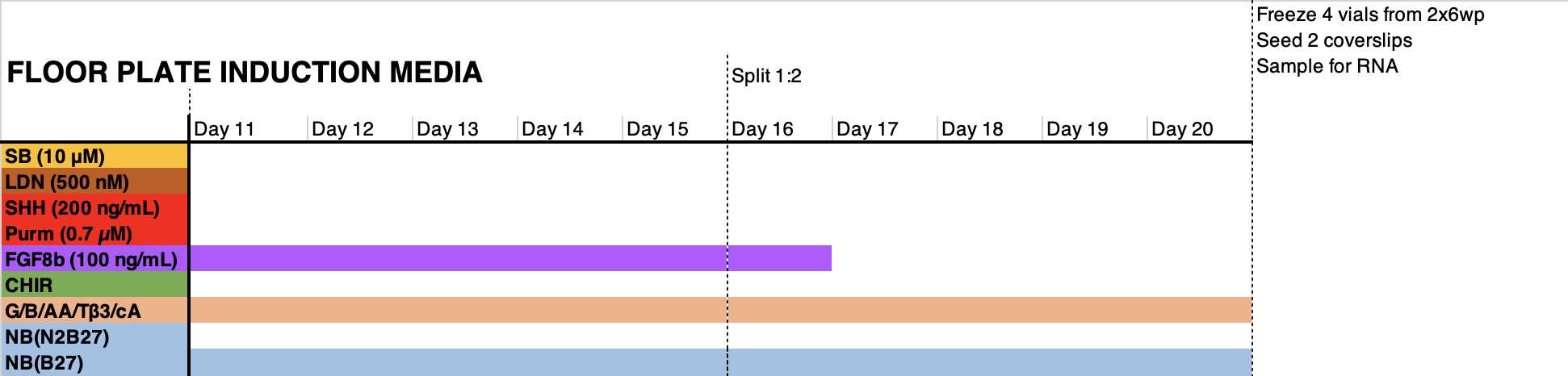
Terminal differentiation
Day 21: thaw 1 cryovial in a Matrigel-coated well of a 12wp in NB(B27) medium in the presence of RI.
Day 22: refresh the medium with Terminal Differentiation medium (see materials section).
Day 24: preincubate with RI from 2h to O/N. Disaggregate cells using accutase and reseed them onto murine laminin or human Laminin521-coated wells (with or without coverslips) at the desired density (generally, 75,000-150,000 cells/cm2 works well).
Quality control
Immunofluorescence analysis
The different progenitors should express markers specific to the ventricular and subventricular zones of the corresponding brain regions:
- Cortical progenitors: FOXG1/OTX2/PAX6/TBR2
- Striatal progenitors: FOXG1/OTX2/GSX2/DLX2/SIX3
- Mesencephalic floor plate progenitors: FOXA2/LMX1A/OTX2/EN1
RT-qPCR
It is advisable to test a battery of markers that are specific to the developing brain regions of interest but also to include markers specific to the pluripotent state as well as markers specific to neighboring regions to rule out contaminations.
With respect to the latter, we regularly include markers specific to the lateral (GSX2 and DLX2) and medial (NKX2.1 and DLX2) ganglionic eminences when checking cortical progenitors. We include markers specific to the developing cortex and to the medial ganglionic eminence (NKX2.1) when checking lateral ganglionic eminence (striatal) progenitors. And finally, we include markers specific to regions anterior (BARHL1), posterior (HOXA2), or dorsal (NKX6-1) to the mesencephalic floor plate when checking mesencephalic floor plate progenitors.

